3PMR
 
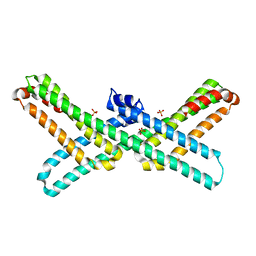 | | Crystal Structure of E2 domain of Human Amyloid Precursor-Like Protein 1 | | Descriptor: | Amyloid-like protein 1, PHOSPHATE ION | | Authors: | Lee, S, Xue, Y, Hu, J, Wang, Y, Liu, X, Demeler, B, Ha, Y. | | Deposit date: | 2010-11-17 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
7MR3
 
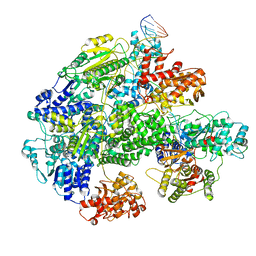 | | Cryo-EM structure of RecBCD-DNA complex with docked RecBNuc and stabilized RecD | | Descriptor: | DNA (60-MER), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Hao, L, Zhang, R, Lohman, T.M. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Heterogeneity in E. coli RecBCD Helicase-DNA Binding and Base Pair Melting.
J.Mol.Biol., 433, 2021
|
|
3V9K
 
 | |
3TRP
 
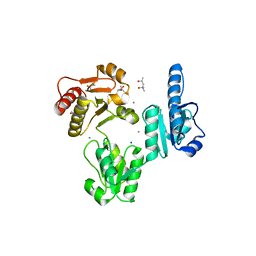 | | Crystal structure of recombinant rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8817 Å) | | Cite: | High-capacity Ca2+ Binding of Human Skeletal Calsequestrin.
J.Biol.Chem., 287, 2012
|
|
7MR2
 
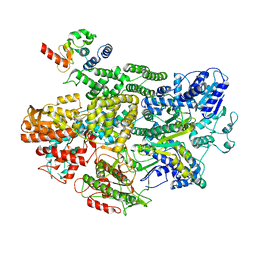 | |
7MR4
 
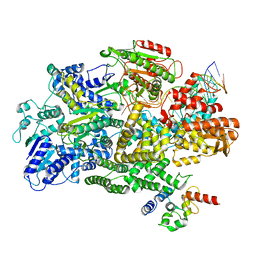 | | Cryo-EM structure of RecBCD-DNA complex with undocked RecBNuc and flexible RecD | | Descriptor: | DNA (60-MER), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Hao, L, Zhang, R, Lohman, T.M. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Heterogeneity in E. coli RecBCD Helicase-DNA Binding and Base Pair Melting.
J.Mol.Biol., 433, 2021
|
|
1VEV
 
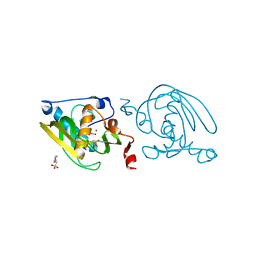 | | Crystal structure of peptide deformylase from Leptospira Interrogans (LiPDF) at pH6.5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FORMIC ACID, Peptide deformylase, ... | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-04-06 | | Release date: | 2005-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Novel conformational states of peptide deformylase from pathogenic bacterium Leptospira interrogans: implications for population shift
J.Biol.Chem., 280, 2005
|
|
7MWC
 
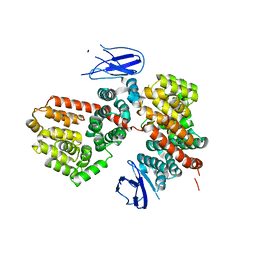 | | ERAP1 binds peptide C-terminus of a LPF sequence (AAAAFKARKF) | | Descriptor: | Endoplasmic reticulum aminopeptidase 1,LPF sequence | | Authors: | Guo, H.C, Sui, L. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ERAP1 binds peptide C-termini of different sequences and/or lengths by a common recognition mechanism.
Immunobiology, 226, 2021
|
|
3POP
 
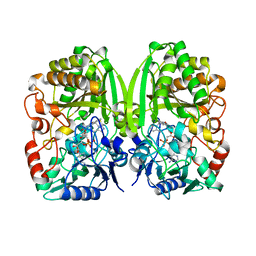 | | The crystal structure of GilR, an oxidoreductase that catalyzes the terminal step of gilvocarcin biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GilR oxidase | | Authors: | Noinaj, N, Bosserman, M.A, Schickli, M.A, Kharel, M.K, Rohr, J, Buchanan, S.K. | | Deposit date: | 2010-11-23 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | The Crystal Structure and Mechanism of an Unusual Oxidoreductase, GilR, Involved in Gilvocarcin V Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
7MF4
 
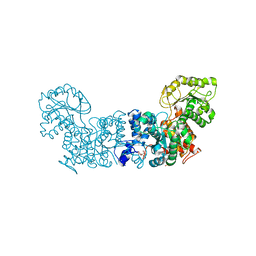 | |
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
7M5I
 
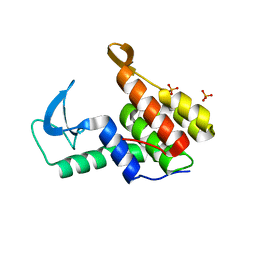 | | Endolysin from Escherichia coli O157:H7 phage FAHEc1 | | Descriptor: | Endolysin, PHOSPHATE ION | | Authors: | Love, M.J, Coombes, D, Billington, C, Dobson, R.C.J. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Molecular Basis for Escherichia coli O157:H7 Phage FAHEc1 Endolysin Function and Protein Engineering to Increase Thermal Stability.
Viruses, 13, 2021
|
|
5DL0
 
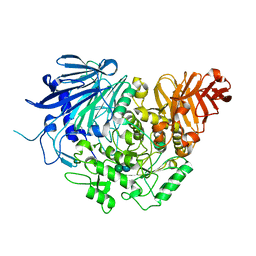 | | Crystal structure of glucosidase II alpha subunit (Glc1Man2-bound from) | | Descriptor: | Alpha glucosidase-like protein, alpha-D-glucopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
7MWB
 
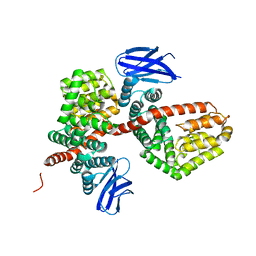 | | ERAP1 binds peptide C-terminus of a SPF sequence (FKARKF) | | Descriptor: | Endoplasmic reticulum aminopeptidase 1,SPF Sequence | | Authors: | Guo, H.C, Sui, L. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | ERAP1 binds peptide C-termini of different sequences and/or lengths by a common recognition mechanism.
Immunobiology, 226, 2021
|
|
5T31
 
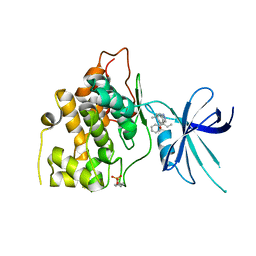 | | Exploiting an Asp-Glu switch in Glycogen Synthase Kinase 3 to design paralog selective inhibitors for use in acute myeloid leukemia | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Stein, A.J, Holson, E.B, Wagner, F.F, Cambell, A.J. | | Deposit date: | 2016-08-24 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5EDD
 
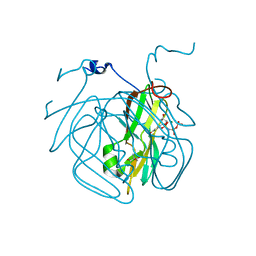 | | Crystal structure of Mycobacterium tuberculosis dUTPase R140K, H145W mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Nagy, G.N, Leveles, I, Lopata, A, Harmat, V, Toth, J, Vertessy, G.B. | | Deposit date: | 2015-10-21 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Characterization of Arginine Fingers: Identification of an Arginine Finger for the Pyrophosphatase dUTPases.
J. Am. Chem. Soc., 138, 2016
|
|
5DL9
 
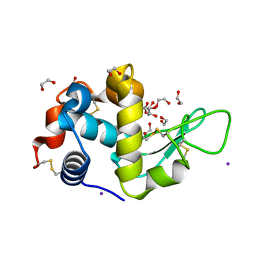 | | Structure of Tetragonal Lysozyme in complex with Iodine solved by UWO Students | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IODIDE ION, ... | | Authors: | Bednarski, R, Cirricione, N, Greco, A, Hodgson, R, Kent, S, McGowan, J, Notherm, B, Patt, M, Vue, L, Bianchetti, C.M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of Tetragonal Lysozyme in complex with Iodine solved by UWO Students
To Be Published
|
|
1A4O
 
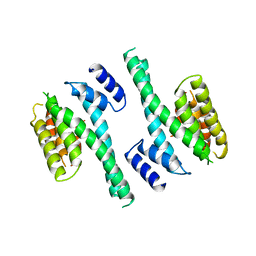 | | 14-3-3 PROTEIN ZETA ISOFORM | | Descriptor: | 14-3-3 PROTEIN ZETA | | Authors: | Liu, D, Bienkowska, J, Petosa, C, Collier, R.J, Fu, H, Liddington, R.C. | | Deposit date: | 1998-02-01 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the zeta isoform of the 14-3-3 protein.
Nature, 376, 1995
|
|
7M8E
 
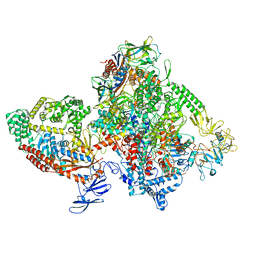 | | E.coli RNAP-RapA elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, W, Liu, B. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for activation of Swi2/Snf2 ATPase RapA by RNA polymerase.
Nucleic Acids Res., 49, 2021
|
|
3U9P
 
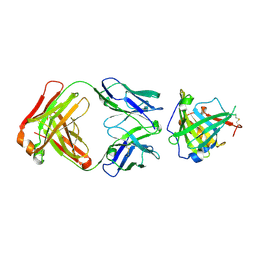 | | Crystal Structure of Murine Siderocalin in Complex with an Fab Fragment | | Descriptor: | Monoclonal Fab Fragment Heavy Chain, Monoclonal Fab Fragment Light Chain, Neutrophil gelatinase-associated lipocalin | | Authors: | Correnti, C, Strong, R.K. | | Deposit date: | 2011-10-19 | | Release date: | 2013-05-01 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Siderocalin/Lcn2/NGAL/24p3 does not drive apoptosis through gentisic acid mediated iron withdrawal in hematopoietic cell lines.
Plos One, 7, 2012
|
|
3V9L
 
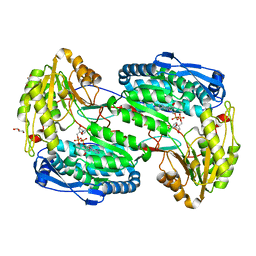 | |
3PUM
 
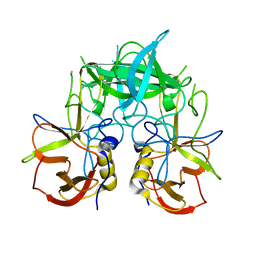 | | Crystal structure of P domain dimer of Norovirus VA207 | | Descriptor: | Capsid | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
3CO1
 
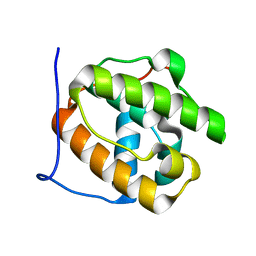 | |
1A17
 
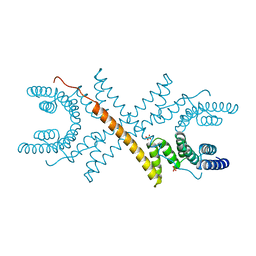 | | TETRATRICOPEPTIDE REPEATS OF PROTEIN PHOSPHATASE 5 | | Descriptor: | SERINE/THREONINE PROTEIN PHOSPHATASE 5, SULFATE ION | | Authors: | Das, A.K, Cohen, P.T.W, Barford, D. | | Deposit date: | 1997-12-23 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions.
EMBO J., 17, 1998
|
|
1DOQ
 
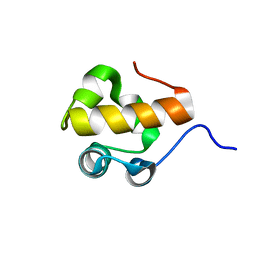 | |
