1LLM
 
 | |
3L7K
 
 | | Structure of the Wall Teichoic Acid Polymerase TagF, H444N + CDPG (15 minute soak) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Strynadka, N.C.J, Lovering, A.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5ZIH
 
 | | Crystal structure of the red light-activated channelrhodopsin Chrimson. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sensory opsin A,Chrimson | | Authors: | Oda, K, Vierock, J, Oishi, S, Taniguchi, R, Yamashita, K, Nishizawa, T, Hegemann, P, Nureki, O. | | Deposit date: | 2018-03-15 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the red light-activated channelrhodopsin Chrimson.
Nat Commun, 9, 2018
|
|
1LCA
 
 | | LACTOBACILLUS CASEI THYMIDYLATE SYNTHASE TERNARY COMPLEX WITH DUMP AND CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-06-22 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined structures of substrate-bound and phosphate-bound thymidylate synthase from Lactobacillus casei.
J.Mol.Biol., 232, 1993
|
|
5ZJK
 
 | | Structure of myroilysin | | Descriptor: | Myroilysin, PHOSPHATE ION, ZINC ION | | Authors: | Li, W.D, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2018-03-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of mature myroilysin and implication for its activation mechanism.
Int.J.Biol.Macromol., 2019
|
|
1LBG
 
 | | LACTOSE OPERON REPRESSOR BOUND TO 21-BASE PAIR SYMMETRIC OPERATOR DNA, ALPHA CARBONS ONLY | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), PROTEIN (LACTOSE OPERON REPRESSOR) | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-01-03 | | Release date: | 1996-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
5Z0B
 
 | |
1LFD
 
 | | CRYSTAL STRUCTURE OF THE ACTIVE RAS PROTEIN COMPLEXED WITH THE RAS-INTERACTING DOMAIN OF RALGDS | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RALGDS, ... | | Authors: | Huang, L, Hofer, F, Martin, G.S, Kim, S.-H. | | Deposit date: | 1998-04-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction of Ras with RalGDS.
Nat.Struct.Biol., 5, 1998
|
|
5Z2L
 
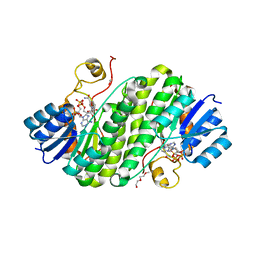 | | Crystal structure of BdcA in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Cyclic-di-GMP-binding biofilm dispersal mediator protein, ... | | Authors: | Yang, W.S, Hou, Y.J, Li, D.F, Wang, D.C. | | Deposit date: | 2018-01-03 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A potential substrate binding pocket of BdcA plays a critical role in NADPH recognition and biofilm dispersal
Biochem. Biophys. Res. Commun., 497, 2018
|
|
1LG1
 
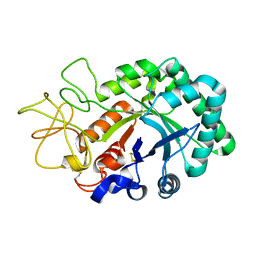 | |
1LJJ
 
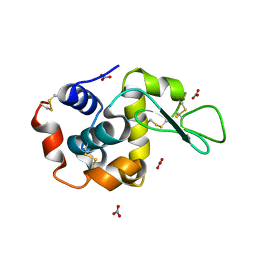 | |
5Z2P
 
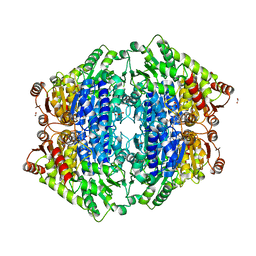 | | ThDP-Mn2+ complex of R413K variant of EcMenD soaked with 2-ketoglutarate for 5 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, FORMIC ACID, ... | | Authors: | Qin, M.M, Guo, Z.H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two active site arginines are critical determinants of substrate binding and catalysis in MenD: a thiamine-dependent enzyme in menaquinone biosynthesis.
Biochem. J., 475, 2018
|
|
5ZBO
 
 | | Cryo-EM structure of PCV2 VLPs | | Descriptor: | Capsid protein | | Authors: | Mo, X, Yuan, A.Y. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structural roles of PCV2 capsid protein N-terminus in PCV2 particle assembly and identification of PCV2 type-specific neutralizing epitope.
PLoS Pathog., 15, 2019
|
|
1LKM
 
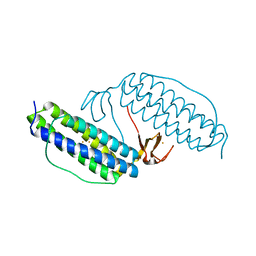 | | Crystal structure of Desulfovibrio vulgaris rubrerythrin all-iron(III) form | | Descriptor: | FE (III) ION, Rubrerythrin all-iron(III) form | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.J, Rose, J, Wang, B.C. | | Deposit date: | 2002-04-25 | | Release date: | 2002-09-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | X-ray Crystal Structures of Reduced Rubrerythrin and its Azide Adduct: A Structure-Based Mechanism for a Non-Heme DiIron Peroxidase
J.Am.Chem.Soc., 124, 2002
|
|
1LL1
 
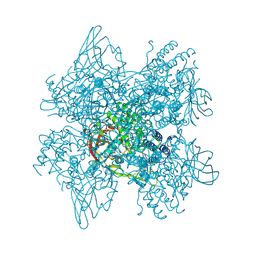 | | HYDROXO BRIDGE MET FORM HEMOCYANIN FROM LIMULUS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, METCYANIN II | | Authors: | Liu, S, Magnus, K. | | Deposit date: | 1997-03-17 | | Release date: | 1997-08-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic Studies of Hydroxo, Nitrosyl, Azido and Fluro met Form Hemocyanin Subunit II from Limulus
To be Published
|
|
1LLI
 
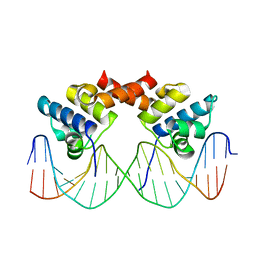 | | THE CRYSTAL STRUCTURE OF A MUTANT PROTEIN WITH ALTERED BUT IMPROVED HYDROPHOBIC CORE PACKING | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Lim, W.A, Hodel, A, Sauer, R.T, Richards, F.M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a mutant protein with altered but improved hydrophobic core packing.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1LAJ
 
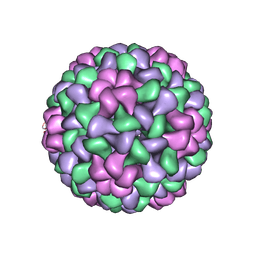 | | The Structure of Tomato Aspermy Virus by X-Ray Crystallography | | Descriptor: | 5'-R(*AP*AP*A)-3', MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lucas, R.W, Larson, S.B, Canady, M.A, McPherson, A. | | Deposit date: | 2002-03-28 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structure of Tomato Aspermy Virus by X-Ray Crystallography
J.STRUCT.BIOL., 139, 2002
|
|
1LBH
 
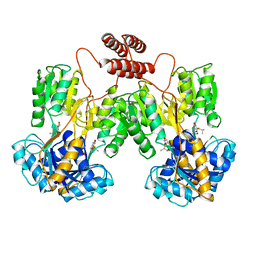 | | INTACT LACTOSE OPERON REPRESSOR WITH GRATUITOUS INDUCER IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, INTACT LACTOSE OPERON REPRESSOR WITH GRATUITOUS INDUCER IPTG | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-02-17 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
1LAM
 
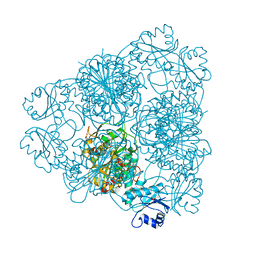 | | LEUCINE AMINOPEPTIDASE (UNLIGATED) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CARBONATE ION, LEUCINE AMINOPEPTIDASE, ... | | Authors: | Straeter, N, Lipscomb, W.N. | | Deposit date: | 1995-08-11 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two-metal ion mechanism of bovine lens leucine aminopeptidase: active site solvent structure and binding mode of L-leucinal, a gem-diolate transition state analogue, by X-ray crystallography.
Biochemistry, 34, 1995
|
|
1LD7
 
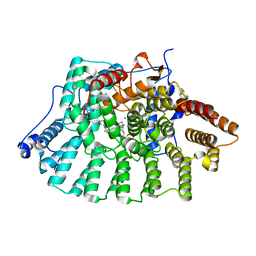 | | Co-crystal structure of Human Farnesyltransferase with farnesyldiphosphate and inhibitor compound 66 | | Descriptor: | (20S)-19,20,22,23-TETRAHYDRO-19-OXO-5H,21H-18,20-ETHANO-12,14-ETHENO-6,10-METHENOBENZ[D]IMIDAZO[4,3-L][1,6,9,13]OXATRIA ZACYCLONOADECOSINE-9-CARBONITRILE, FARNESYL DIPHOSPHATE, ZINC ION, ... | | Authors: | Taylor, J.S, Terry, K.L, Beese, L.S. | | Deposit date: | 2002-04-08 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-Aminopyrrolidinone farnesyltransferase inhibitors: design of macrocyclic compounds with improved pharmacokinetics and excellent cell potency.
J.Med.Chem., 45, 2002
|
|
1LG2
 
 | |
1LJF
 
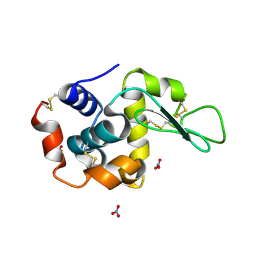 | |
1LJT
 
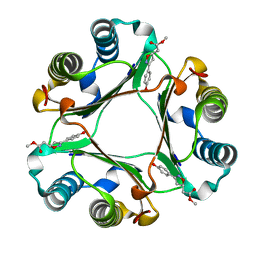 | | Crystal Structure of Macrophage Migration Inhibitory Factor complexed with (S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole-acetic acid methyl ester (ISO-1) | | Descriptor: | 3-(4-HYDROXYPHENYL)-4,5-DIHYDRO-5-ISOXAZOLE-ACETIC ACID METHYL ESTER, Macrophage migration inhibitory factor | | Authors: | Lubetsky, J.B, Dios, A, Han, J, Aljabari, B, Ruzsicska, B, Mitchell, R, Lolis, E, Al-Abed, Y. | | Deposit date: | 2002-04-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Tautomerase Active Site of Macrophage Migration Inhibitory factor is a Potential Target for Discovery of Novel Anti-inflammatory Agents
J.Biol.Chem., 277, 2002
|
|
1LKF
 
 | | LEUKOCIDIN F (HLGB) FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | LEUKOCIDIN F SUBUNIT | | Authors: | Olson, R, Nariya, H, Yokota, K, Kamio, Y, Gouaux, J.E. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel.
Nat.Struct.Biol., 6, 1999
|
|
5ZAB
 
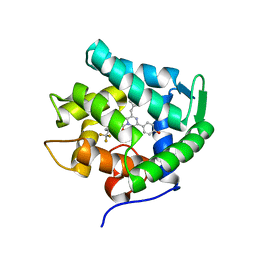 | | Crystal structure of cf3-aequorin | | Descriptor: | (2S)-8-benzyl-2-hydroperoxy-6-(4-hydroxyphenyl)-2-{[4-(trifluoromethyl)phenyl]methyl}imidazo[1,2-a]pyrazin-3(2H)-one, Aequorin-2 | | Authors: | Inouye, S, Tomabechi, Y, Sekine, S.I, Shirouzu, M, Hosoya, T. | | Deposit date: | 2018-02-07 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Slow luminescence kinetics of semi-synthetic aequorin: expression, purification and structure determination of cf3-aequorin.
J. Biochem., 164, 2018
|
|
