1MJQ
 
 | |
5LZU
 
 | | Structure of the mammalian ribosomal termination complex with accommodated eRF1 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5WAI
 
 | | Crystal Structure of a Suz12-Rbbp4-Jarid2-Aebp2 Heterotetrameric Complex | | Descriptor: | Histone-binding protein RBBP4, Jumonji, AT-rich interactive domain 2, ... | | Authors: | Chen, S, Jiao, L, Liu, X. | | Deposit date: | 2017-06-26 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding.
Mol. Cell, 69, 2018
|
|
1MJP
 
 | |
5LMK
 
 | | Structure of phopsho-CDK2-cyclin A in complex with an ATP-competitive inhibitor | | Descriptor: | 4-[4-[3-bromanyl-7-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-5-yl]phenyl]benzamide, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Explicit treatment of active-site waters enhances quantum mechanical/implicit solvent scoring: Inhibition of CDK2 by new pyrazolo[1,5-a]pyrimidines.
Eur J Med Chem, 126, 2016
|
|
4UY8
 
 | | Molecular basis for the ribosome functioning as a L-tryptophan sensor - Cryo-EM structure of a TnaC stalled E.coli ribosome | | Descriptor: | 50S RIBOSOMAL PROTEIN L10, 50S RIBOSOMAL PROTEIN L11, 50S RIBOSOMAL PROTEIN L13, ... | | Authors: | Bischoff, L, Berninghausen, O, Beckmann, R. | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-29 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular Basis for the Ribosome Functioning as an L-Tryptophan Sensor.
Cell Rep., 9, 2014
|
|
6OQI
 
 | | CDK2 in complex with Cpd14 (5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)-N-(5-(4-methylpiperazin-1-yl)pyridin-2-yl)pyrimidin-2-amine) | | Descriptor: | 5-fluoro-N-[5-(4-methylpiperazin-1-yl)pyridin-2-yl]-4-[(4S)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Murray, J.M. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4V7J
 
 | | Structure of RelE nuclease bound to the 70S ribosome (precleavage state) | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | Deposit date: | 2009-11-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
4W2I
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with negamycin, mRNA and three deacylated tRNAs in the A, P and E sites | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Szal, T, Jiang, F, Gupta, P, Matsuda, R, Shiozuka, M, Steitz, T.A, Vazquez-Laslop, N, Mankin, A.S. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Negamycin Interferes with Decoding and Translocation by Simultaneous Interaction with rRNA and tRNA.
Mol.Cell, 56, 2014
|
|
8IPD
 
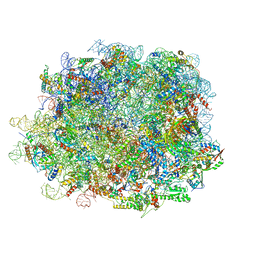 | | human nuclear pre-60S ribosomal particle - State C | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-03-14 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
3SQQ
 
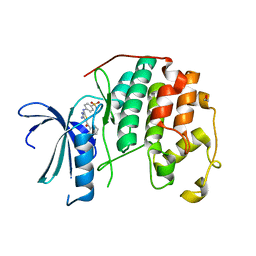 | | CDK2 in complex with inhibitor RC-3-96 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(2-methylbenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-07-06 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3SW4
 
 | |
6ONO
 
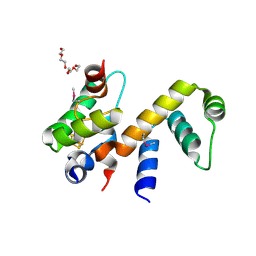 | | Complex structure of WhiB1 and region 4 of SigA in C2221 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | Authors: | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-11-27 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
6ONU
 
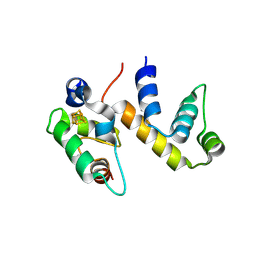 | | Complex structure of WhiB1 and region 4 of SigA in P21 space group. | | Descriptor: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | Authors: | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
7OIZ
 
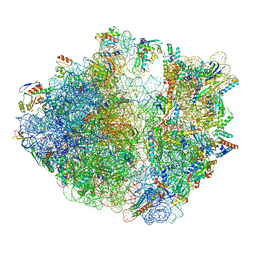 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Kudva, R, Becker, T, Berninghausen, O, Heijne, G, Cheng, J, Beckmann, R. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
7OJ0
 
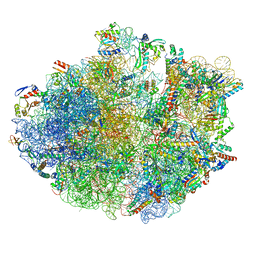 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide and RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Kudva, R, Becker, T, Berninghausen, O, Heijne, G, Cheng, J, Beckmann, R. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
3T3Y
 
 | |
1VQJ
 
 | | GENE V PROTEIN MUTANT WITH VAL 35 REPLACED BY ILE 35 (V35I) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1VQH
 
 | |
3T4H
 
 | | Crystal Structure of AlkB in complex with Fe(III) and N-Oxalyl-S-(3-nitrobenzyl)-L-cysteine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, FE (III) ION, ... | | Authors: | Ma, J, Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-07-26 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dynamic combinatorial mass spectrometry leads to inhibitors of a 2-oxoglutarate-dependent nucleic Acid demethylase.
J.Med.Chem., 55, 2012
|
|
1VQA
 
 | |
1VYW
 
 | | Structure of CDK2/Cyclin A with PNU-292137 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, N-(3-CYCLOPROPYL-1H-PYRAZOL-5-YL)-2-(2-NAPHTHYL)ACETAMIDE, ... | | Authors: | Pevarello, P, Brasca, M.G, Amici, R, Orsini, P, Traquandi, G, Corti, L, Piutti, C, Sansonna, P, Villa, M, Pierce, B.S, Pulici, M, Giordano, P, Martina, K, Fritzen, E.L, Nugent, R.A, Casale, E, Cameron, A, Ciomei, M, Roletto, F, Isacchi, A, Fogliatto, G, Pesenti, E, Pastori, W, Marsiglio, A, Leach, K.L, Clare, P.M, Fiorentini, F, Varasi, M, Vulpetti, A, Warpehoski, M.A. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2/Cyclin a as Antitumor Agents. Part 1. Lead Finding
J.Med.Chem., 47, 2004
|
|
7OLE
 
 | | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2, ... | | Authors: | Pal, M, Llorca, O, Pearl, L. | | Deposit date: | 2021-05-19 | | Release date: | 2021-07-07 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of the TELO2-TTI1-TTI2 complex and its function in TOR recruitment to the R2TP chaperone.
Cell Rep, 36, 2021
|
|
8IR1
 
 | | human nuclear pre-60S ribosomal particle - State A | | Descriptor: | 28S rRNA, 5.8S rRMA, 5S RNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-03-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
7P6X
 
 | | Cryo-Em structure of the hexameric RUVBL1-RUVBL2 in complex with ZNHIT2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Llorca, O, Serna, M, Fernandez-Leiro, R. | | Deposit date: | 2021-07-18 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | CryoEM of RUVBL1-RUVBL2-ZNHIT2, a complex that interacts with pre-mRNA-processing-splicing factor 8.
Nucleic Acids Res., 50, 2022
|
|
