1SGU
 
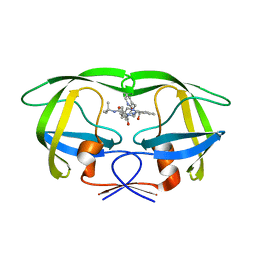 | | Comparing the Accumulation of Active Site and Non-active Site Mutations in the HIV-1 Protease | | Descriptor: | N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, POL polyprotein | | Authors: | Clemente, J.C, Moose, R.E, Hemrajani, R, Govindasamy, L, Reutzel, R, Mckenna, R, Abandje-McKenna, M, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2004-02-24 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparing the Accumulation of Active- and Nonactive-Site Mutations in the HIV-1 Protease.
Biochemistry, 43, 2004
|
|
1XQL
 
 | | Effect of a Y265F Mutant on the Transamination Based Cycloserine Inactivation of Alanine Racemase | | Descriptor: | (5-HYDROXY-4-{[(3-HYDROXYISOXAZOL-4-YL)AMINO]METHYL}-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, (R)-4-AMINO-ISOXAZOLIDIN-3-ONE, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Fenn, T.D, Holyoak, T, Stamper, G.F, Ringe, D. | | Deposit date: | 2004-10-12 | | Release date: | 2005-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of a Y265F Mutant on the Transamination-Based Cycloserine Inactivation of Alanine Racemase
Biochemistry, 44, 2005
|
|
1XRN
 
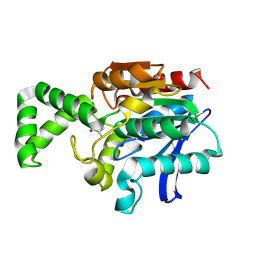 | | Crystal structure of active site F1-mutant E213Q soaked with peptide Phe-Ala | | Descriptor: | ALANINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1S1S
 
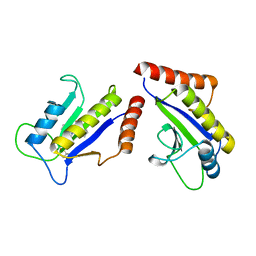 | | Crystal Structure of ZipA in complex with indoloquinolizin 10b | | Descriptor: | Cell division protein zipA, N-{3-[(12bS)-7-oxo-1,3,4,6,7,12b-hexahydroindolo[2,3-a]quinolizin-12(2H)-yl]propyl}propane-2-sulfonamide | | Authors: | Jennings, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenny, C.H, Moghazeh, S.L, Petersen, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | Deposit date: | 2004-01-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
2BMW
 
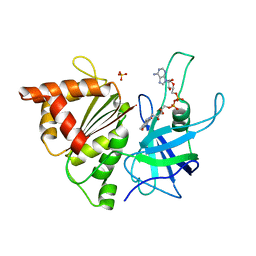 | | Ferredoxin: NADP+ Reductase Mutant With Thr 155 Replaced By Gly, Ala 160 Replaced By Thr, Leu 263 Replaced By Pro, Arg 264 Replaced By Pro and Gly 265 Replaced by Pro (T155G-A160T-L263P-R264P-G265P) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Martinez-Julvez, M, Hermoso, J.A, Perez-Dorado, I, Medina, M, Tejero, J, Gomez-Moreno, C. | | Deposit date: | 2005-03-16 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein Motifs Involved in Coenzyme Interaction and Enzymatic Efficiency in Anabaena Ferredoxin-Nadp+ Reductase.
Biochemistry, 48, 2009
|
|
2BUI
 
 | | E. COLI BETA-KETOACYL (ACYL CARRIER PROTEIN) SYNTHASE I IN COMPLEX WITH OCTANOIC ACID, 120K | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE I, AMMONIUM ION, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Olsen, J.G, Von Wettstein-Knowles, P, Henriksen, A. | | Deposit date: | 2005-06-13 | | Release date: | 2005-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fatty acid synthesis. Role of active site histidines and lysine in Cys-His-His-type beta-ketoacyl-acyl carrier protein synthases.
FEBS J., 273, 2006
|
|
1XZC
 
 | |
1SLX
 
 | |
2BQV
 
 | | HIV-1 protease in complex with inhibitor AHA455 | | Descriptor: | 6-AMINO HEXANOIC ACID, HIV-1 PROTEASE | | Authors: | Unge, T, Ekegren, J.K, Schenk, H.V, Zreik Safa, M, Wallberg, H, Samuelsson, B, Hallberg, A. | | Deposit date: | 2005-04-28 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A New Class of HIV-1 Protease Inhibitors Containing a Tertiary Alcohol in the Transition-State Mimicking Scaffold.
J.Med.Chem., 48, 2005
|
|
1XTK
 
 | | structure of DECD to DEAD mutation of human UAP56 | | Descriptor: | BETA-MERCAPTOETHANOL, Probable ATP-dependent RNA helicase p47 | | Authors: | Shi, H, Cordin, O, Minder, C.M, Linder, P, Xu, R.M. | | Deposit date: | 2004-10-22 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human ATP-dependent splicing and export factor UAP56
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SM4
 
 | | Crystal Structure Analysis of the Ferredoxin-NADP+ Reductase from Paprika | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, chloroplast ferredoxin-NADP+ oxidoreductase | | Authors: | Dorowski, A, Hofmann, A, Steegborn, C, Boicu, M, Huber, R. | | Deposit date: | 2004-03-08 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of paprika ferredoxin-NADP+ reductase. Implications for the electron transfer pathway.
J.Biol.Chem., 276, 2001
|
|
1SME
 
 | | PLASMEPSIN II, A HEMOGLOBIN-DEGRADING ENZYME FROM PLASMODIUM FALCIPARUM, IN COMPLEX WITH PEPSTATIN A | | Descriptor: | PLASMEPSIN II, Pepstatin | | Authors: | Silva, A.M, Lee, A.Y, Gulnik, S.V, Goldberg, D.E, Erickson, J.W. | | Deposit date: | 1996-06-11 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparum.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
2BQ6
 
 | | Crystal structure of factor Xa in complex with 21 | | Descriptor: | 1-{[5-(5-CHLORO-2-THIENYL)ISOXAZOL-3-YL]METHYL}-3-CYANO-N-(1-ISOPROPYLPIPERIDIN-4-YL)-7-METHYL-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-27 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
1XF1
 
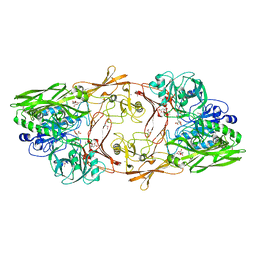 | | Structure of C5a peptidase- a key virulence factor from Streptococcus | | Descriptor: | ACETATE ION, C5a peptidase, CALCIUM ION, ... | | Authors: | Brown, C.K, Gu, Z.Y, Cleary, P.P, Matsuka, Y, Olmstead, S, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2004-09-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the streptococcal cell wall C5a peptidase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1S39
 
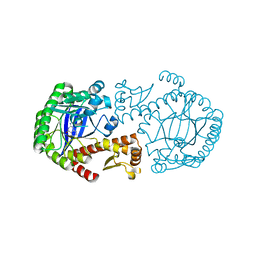 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2-aminoquinazolin-4(3H)-one | | Descriptor: | 2-AMINOQUINAZOLIN-4(3H)-ONE, ZINC ION, tRNA guanine transglycosylase | | Authors: | Brenk, R, Meyer, E.A, Reuter, K, Garcia, G.A, Stubbs, M.T, Klebe, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis and In Vitro Evaluation of 2-Aminoquinazolin-4(3H)-one-Based Inhibitors for tRNA-Guanine Transglycosylase (TGT)
HELV.CHIM.ACTA, 87, 2004
|
|
2BQ3
 
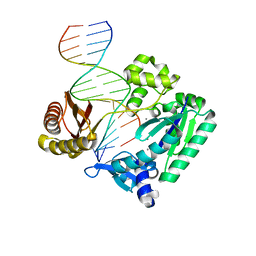 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*A)-3', 5'-D(*TP*CP*AP*TP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', CALCIUM ION, ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-27 | | Release date: | 2005-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
1XUH
 
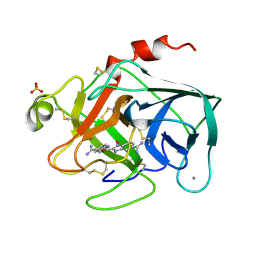 | | TRYPSIN-KETO-BABIM-CO+2, PH 8.2 | | Descriptor: | BIS(5-AMIDINO-2-BENZIMIDAZOLYL)METHANONE, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Katz, B.A, Clark, J.M, Finer-Moore, J.S, Jenkins, T.E, Johnson, C.R, Rose, M.J, Luong, C, Moore, W.R, Stroud, R.M. | | Deposit date: | 1997-10-10 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of potent selective zinc-mediated serine protease inhibitors.
Nature, 391, 1998
|
|
2BXN
 
 | | Human serum albumin complexed with myristate and iodipamide | | Descriptor: | 3-[5-[(3-CARBOXY-2,4,6-TRIIODO-PHENYL)CARBAMOYL]PENTANOYLAMINO]-2,4,6-TRIIODO-BENZOIC ACID, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
1XET
 
 | | Crystal structure of stilbene synthase from Pinus sylvestris, complexed with methylmalonyl CoA | | Descriptor: | 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, Dihydropinosylvin synthase, METHYLMALONYL-COENZYME A | | Authors: | Ng, S.H, Chirgadze, D, Spiteller, D, Li, T.L, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2004-09-12 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of stilbene synthase from Pinus sylvestris, complexed with methylmalonyl CoA
To be Published
|
|
1RT2
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Willcox, B, Jones, Y, Ross, C, Stammers, D, Stuart, D. | | Deposit date: | 1996-03-16 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors.
J.Med.Chem., 39, 1996
|
|
2BPY
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | HIV-1 PROTEASE, N-[2(S)-CYCLOPENTYL-1(R)-HYDROXY-3(R)METHYL]-5-[(2(S)-TERTIARY-BUTYLAMINO-CARBONYL)-4-(N1-(2)-(N-METHYLPIPERAZINYL)-3-CHLORO-PYRAZINYL-5-CARBONYL)-PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYL-PENTANAMIDE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BMH
 
 | |
2BPZ
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | HIV-1 PROTEASE, N-[2(S)-CYCLOPENTYL-1(R)-HYDROXY-3(R)METHYL]-5-[(2(S)-TERTIARY-BUTYLAMINO-CARBONYL)-4-(N1-(2)-(N-METHYLPIPERAZINYL)-3-CHLORO-PYRAZINYL-5-CARBONYL)-PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYL-PENTANAMIDE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPW
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | 1-[2-HYDROXY-4-(2-HYDROXY-5-METHYL-CYCLOPENTYLCARBAMOYL)5-PHENYL-PENTYL]-4-(3-PYRIDIN-3-YL-PROPIONYL)-PIPERAZINE-2-CARB OXYLIC ACID TERT-BUTYLAMIDE, HIV-1 PROTEASE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1RPA
 
 | |
