3FCS
 
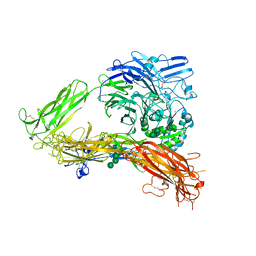 | | Structure of complete ectodomain of integrin aIIBb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
5VHZ
 
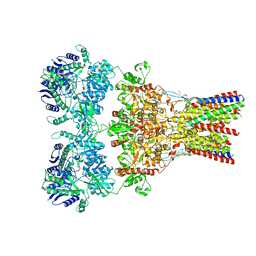 | | GluA2-2xGSG1L bound to L-Quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2,Germ cell-specific gene 1-like protein | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-03 | | Last modified: | 2017-11-08 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes.
Neuron, 94, 2017
|
|
6NEE
 
 | |
3QHN
 
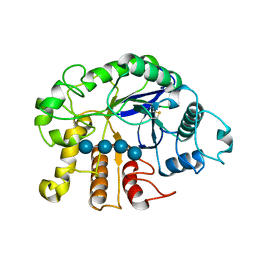 | | Crystal analysis of the complex structure, E201A-cellotetraose, of endocellulase from pyrococcus horikoshii | | Descriptor: | 458aa long hypothetical endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional analysis of hyperthermophilic endocellulase from Pyrococcus horikoshii by crystallographic snapshots
Biochem.J., 437, 2011
|
|
2O5Y
 
 | |
2KKJ
 
 | |
1FT7
 
 | | AAP COMPLEXED WITH L-LEUCINEPHOSPHONIC ACID | | Descriptor: | BACTERIAL LEUCYL AMINOPEPTIDASE, LEUCINE PHOSPHONIC ACID, POTASSIUM ION, ... | | Authors: | Stamper, C, Bennett, B, Holz, R, Petsko, G, Ringe, D. | | Deposit date: | 2000-09-11 | | Release date: | 2000-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of the aminopeptidase from Aeromonas proteolytica by L-leucinephosphonic acid. Spectroscopic and crystallographic characterization of the transition state of peptide hydrolysis.
Biochemistry, 40, 2001
|
|
2W3T
 
 | | Chloro complex of the Ni-Form of E.coli deformylase | | Descriptor: | CHLORIDE ION, ETHANOL, NICKEL (II) ION, ... | | Authors: | Ngo, Y.H.T, Palm, G.J, Hinrichs, W. | | Deposit date: | 2008-11-14 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of the Ni(II) Complex of Escherichia Coli Peptide Deformylase and Suggestions on Deformylase Activities Depending on Different Metal(II) Centres.
J.Biol.Inorg.Chem., 15, 2010
|
|
2O5X
 
 | |
8HB2
 
 | |
8HBB
 
 | |
8HAZ
 
 | |
6WHY
 
 | | GluN1b-GluN2B NMDA receptor in complex with GluN1 antagonist L689,560, class 1 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
5EH5
 
 | | human carbonic anhydrase II in complex with ligand | | Descriptor: | 1.7.6 4-methylpyrimidine-2-sulfonamide, Carbonic anhydrase 2, FORMIC ACID, ... | | Authors: | Ren, B. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance, and X-ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
2VWH
 
 | | Haloferax mediterranei glucose dehydrogenase in complex with NADP, Zn and glucose. | | Descriptor: | GLUCOSE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION, ... | | Authors: | Baker, P.J, Britton, K.L, Fisher, M, Esclapez, J, Pire, C, Bonete, M.J, Ferrer, J, Rice, D.W. | | Deposit date: | 2008-06-24 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Active site dynamics in the zinc-dependent medium chain alcohol dehydrogenase superfamily.
Proc. Natl. Acad. Sci. U.S.A., 106, 2009
|
|
6TNT
 
 | | SUMOylated apoAPC/C with repositioned APC2 WHB domain | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Barford, D, Yatskevich, S. | | Deposit date: | 2019-12-10 | | Release date: | 2021-01-13 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular mechanisms of APC/C release from spindle assembly checkpoint inhibition by APC/C SUMOylation.
Cell Rep, 34, 2021
|
|
4Q93
 
 | |
2VWG
 
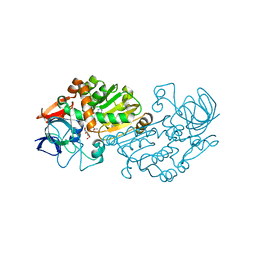 | | Haloferax mediterranei glucose dehydrogenase in complex with NADP, Zn and gluconolactone. | | Descriptor: | D-glucono-1,5-lactone, GLUCOSE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Baker, P.J, Britton, K.L, Fisher, M, Esclapez, J, Pire, C, Bonete, M.J, Ferrer, J, Rice, D.W. | | Deposit date: | 2008-06-24 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site dynamics in the zinc-dependent medium chain alcohol dehydrogenase superfamily.
Proc. Natl. Acad. Sci. U.S.A., 106, 2009
|
|
6QRZ
 
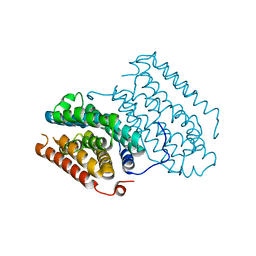 | | Crystal structure of R2-like ligand-binding oxidase from Sulfolobus acidocaldarius solved by 3D micro-crystal electron diffraction | | Descriptor: | FE (III) ION, MANGANESE (III) ION, Ribonucleoside-diphosphate reductase | | Authors: | Xu, H, Lebrette, H, Clabbers, M.T.B, Zhao, J, Griese, J.J, Zou, X, Hogbom, M. | | Deposit date: | 2019-02-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | Solving a new R2lox protein structure by microcrystal electron diffraction.
Sci Adv, 5, 2019
|
|
6WRN
 
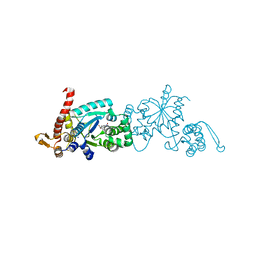 | | Crystal structure of Mj 3-nitro-tyrosine tRNA synthetase (5B) C70A variant bound to 3-nitro-tyrosine | | Descriptor: | META-NITRO-TYROSINE, SODIUM ION, Tyrosine--tRNA ligase | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
8IJC
 
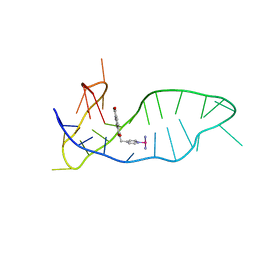 | | NMR solution structure of the 1:1 complex of a platinum(II) ligand L1-transpt covalently bound to a G-quadruplex MYT1L | | Descriptor: | G-quadruplex DNA MYT1L, Pt(NH3)2(2-(pyridin-4-ylmethyl)benzo-[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Mao, Z.-W. | | Deposit date: | 2023-02-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Organic-Platinum Hybrids for Covalent Binding of G-Quadruplexes: Structural Basis and Application to Cancer Immunotherapy.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ICD
 
 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
6WRQ
 
 | | Crystal structure of Mj 3-nitro-tyrosine tRNA synthetase (5B) S158C variant bound to 3-nitro-tyrosine | | Descriptor: | GLYCEROL, META-NITRO-TYROSINE, SODIUM ION, ... | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
8HYE
 
 | | Structure of amino acid dehydrogenase-2752 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alanine dehydrogenase, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-01-06 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two different alanine dehydrogenases from Geobacillus kaustophilus: Their biochemical characteristics and differential expression in vegetative cells and spores.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
1YID
 
 | |
