4RYY
 
 | |
8B4Q
 
 | | Ternary structure of 14-3-3s, ERRg phosphopeptide and dual-reactive compound 5 | | Descriptor: | 14-3-3 protein sigma, 3-fluoranyl-4-methanoyl-~{N}-methyl-~{N}-(2-sulfanylethyl)benzamide, CHLORIDE ION, ... | | Authors: | Somsen, B.A, Ottmann, C. | | Deposit date: | 2022-09-20 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reversible Dual-Covalent Molecular Locking of the 14-3-3/ERR gamma Protein-Protein Interaction as a Molecular Glue Drug Discovery Approach.
J.Am.Chem.Soc., 145, 2023
|
|
6BTL
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor RD117 1-[2-(Piperazin-1-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole | | Descriptor: | 1-[2-(piperazin-1-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bryson, S, De Gasparo, R, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2017-12-06 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Biological Evaluation and X-ray Co-crystal Structures of Cyclohexylpyrrolidine Ligands for Trypanothione Reductase, an Enzyme from the Redox Metabolism of Trypanosoma.
ChemMedChem, 13, 2018
|
|
8IJ7
 
 | |
8DK1
 
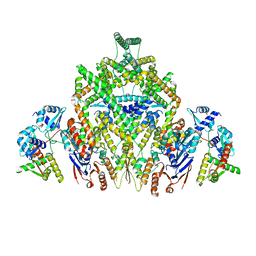 | | CryoEM structure of JetABC (head construct) from Pseudomonas aeruginosa PA14 | | Descriptor: | JetA, JetB, JetC | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
5V72
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with citrate | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Cooper, D.R, Matelska, D, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-17 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
8B6G
 
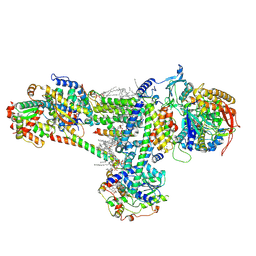 | | Cryo-EM structure of succinate dehydrogenase complex (complex-II) in respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Baradaran, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
5UOG
 
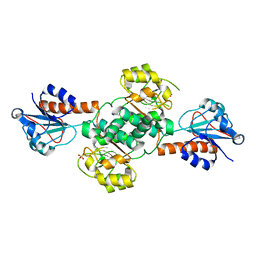 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in apo form | | Descriptor: | NADPH-dependent glyoxylate/hydroxypyruvate reductase, SULFATE ION | | Authors: | Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Cooper, D.R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-01-31 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
8IH7
 
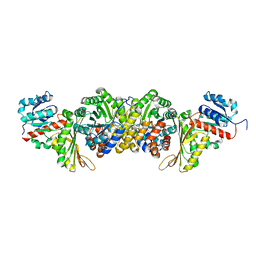 | | AmnG-AmnH complex | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, OXALATE ION, ... | | Authors: | Su, D, Shi, Q.L. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | AmnG-AmnH complex
To Be Published
|
|
1A37
 
 | | 14-3-3 PROTEIN ZETA BOUND TO PS-RAF259 PEPTIDE | | Descriptor: | 14-3-3 PROTEIN ZETA, PS-RAF259 PEPTIDE LSQRQRST(SEP)TPNVHM | | Authors: | Petosa, C, Masters, S.C, Pohl, J, Wang, B, Fu, H, Liddington, R.C. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove.
J.Biol.Chem., 273, 1998
|
|
5UQC
 
 | | Crystal structure of mouse CRMP2 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Dihydropyrimidinase-related protein 2 | | Authors: | Khanna, M, Khanna, R, Perez-Miller, S, Francois-Moutal, L. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A single structurally conserved SUMOylation site in CRMP2 controls NaV1.7 function.
Channels (Austin), 11, 2017
|
|
8GEC
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with nucleotide-free Gs heterotrimer (#13 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
5MGS
 
 | | Human receptor NKR-P1 in deglycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1 | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
8GG1
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #7 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GGM
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #5 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GGX
 
 | | Locally refined cryoEM structure of receptor from beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #16 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
5MHF
 
 | | Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin. | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mannosyl-oligosaccharide glucosidase, ... | | Authors: | Hill, J.C, Caputo, A.T, Roversi, P, Zitzmann, N. | | Deposit date: | 2016-11-24 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Endoplasmic Reticulum alpha-Glucosidase I with a Single-Dose Iminosugar Treatment Protects against Lethal Influenza and Dengue Virus Infections.
J.Med.Chem., 2020
|
|
4AND
 
 | | CRYSTAL FORM II OF THE D93N MUTANT OF NUCLEOSIDE DIPHOSPHATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Georgescauld, F, Moynie, L, Habersetzer, J, Lascu, I, Dautant, A. | | Deposit date: | 2012-03-16 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Intersubunit Ionic Interactions Stabilize the Nucleoside Diphosphate Kinase of Mycobacterium Tuberculosis.
Plos One, 8, 2013
|
|
8GEH
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with nucleotide-free Gs heterotrimer (#18 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GFY
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #4 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
5MJN
 
 | |
8IL5
 
 | |
5MK5
 
 | | Structures of DHBN domain of human BLM helicase | | Descriptor: | Bloom syndrome protein, IODIDE ION, POTASSIUM ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-12-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5NQB
 
 | |
5UTR
 
 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-butyryl-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]butanamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2017-02-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational flexibility of the glycosidase NagZ allows it to bind structurally diverse inhibitors to suppress beta-lactam antibiotic resistance.
Protein Sci., 26, 2017
|
|
