1J12
 
 | | Beta-Amylase from Bacillus cereus var. mycoides in Complex with alpha-EBG | | Descriptor: | 2-[(2S)-oxiran-2-yl]ethyl alpha-D-glucopyranoside, Beta-amylase, CALCIUM ION | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
2FBZ
 
 | | Heme-No complex in a bacterial Nitric Oxide Synthase | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE, ... | | Authors: | Pant, K, Crane, B.R. | | Deposit date: | 2005-12-10 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nitrosyl-heme structures of Bacillus subtilis nitric oxide synthase have implications for understanding substrate oxidation.
Biochemistry, 45, 2006
|
|
2A68
 
 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic rifabutin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2E51
 
 | | Crystal structure of basic winged bean lectin in complex with A blood group disaccharide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2006-12-18 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Generation of blood group specificity: new insights from structural studies on the complexes of A- and B-reactive saccharides with basic winged bean agglutinin.
Proteins, 68, 2007
|
|
3UNP
 
 | | Structure of human SUN2 SUN domain | | Descriptor: | ACETYL GROUP, SUN domain-containing protein 2 | | Authors: | Zhou, Z.C, Greene, M.I. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope
J.Biol.Chem., 287, 2012
|
|
1SJS
 
 | |
1MEZ
 
 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with SAMP, GDP, SO4(2-), and Mg(2+) | | Descriptor: | 2-[9-(3,4-DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-9H-PURIN-6-YLAMINO]-SUCCINIC ACID, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
3UQ3
 
 | | TPR2AB-domain:pHSP90-complex of yeast Sti1 | | Descriptor: | Heat shock protein, Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H, Buchner, J. | | Deposit date: | 2011-11-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
3UXH
 
 | | Design, Synthesis and Biological Evaluation of Potetent Quinoline and Pyrroloquinoline Ammosamide Analogues as Inhibitors of Quinone Reductase 2 | | Descriptor: | 6,8-diamino-7-chloro-1-methyl-2-oxo-1,2-dihydropyrrolo[4,3,2-de]quinoline-4-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Cushman, M, Mesecar, A.D, Fanwick, P.E, Narasimha, R, Jensen, K.C. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design, synthesis, and biological evaluation of potent quinoline and pyrroloquinoline ammosamide analogues as inhibitors of quinone reductase 2.
J.Med.Chem., 55, 2012
|
|
1TJ2
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with acetate | | Descriptor: | ACETATE ION, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
2ASM
 
 | | Structure of Rabbit Actin In Complex With Reidispongiolide A | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Allingham, J.S, Zampella, A, D'Auria, M.V, Rayment, I. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of microfilament destabilizing toxins bound to actin provide insight into toxin design and activity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2ASP
 
 | | Structure of Rabbit Actin In Complex With Reidispongiolide C | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Allingham, J.S, Zampella, A, D'Auria, M.V, Rayment, I. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of microfilament destabilizing toxins bound to actin provide insight into toxin design and activity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3CTS
 
 | |
2DRC
 
 | |
3UZX
 
 | | Crystal structure of 5beta-reductase (AKR1D1) E120H mutant in complex with NADP+ and epiandrosterone | | Descriptor: | (3Beta,5alpha)-3-Hydroxyandrostan-17-one, 3-oxo-5-beta-steroid 4-dehydrogenase, 5-ALPHA-ANDROSTANE-3-BETA,17BETA-DIOL, ... | | Authors: | Chen, M, Christianson, D.W, Penning, T.M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.637 Å) | | Cite: | Conversion of Human Steroid 5beta-Reductase (AKR1D1) into 3β-Hydroxysteroid Dehydrogenase by Single Point Mutation E120H: EXAMPLE OF PERFECT ENZYME ENGINEERING.
J.Biol.Chem., 287, 2012
|
|
1VD3
 
 | | Ribonuclease NT in complex with 2'-UMP | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, RNase NGR3 | | Authors: | Kawano, S, Kakuta, Y, Kimura, M. | | Deposit date: | 2004-03-18 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Nicotiana glutinosa Ribonuclease NT in Complex with Nucleotide Monophosphates
to be published
|
|
2FCX
 
 | | HIV-1 DIS kissing-loop in complex with neamine | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, CHLORIDE ION, HIV-1 DIS RNA, ... | | Authors: | Ennifar, E, Paillart, J.C, Marquet, R, Dumas, P. | | Deposit date: | 2005-12-13 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell.
Nucleic Acids Res., 34, 2006
|
|
3UZZ
 
 | | Crystal structure of 5beta-reductase (AKR1D1) E120H mutant in complex with NADP+ and delta4-androstenedione | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, 4-ANDROSTENE-3-17-DIONE, CHLORIDE ION, ... | | Authors: | Chen, M, Christianson, D.W, Penning, T.M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conversion of Human Steroid 5beta-Reductase (AKR1D1) into 3β-Hydroxysteroid Dehydrogenase by Single Point Mutation E120H: EXAMPLE OF PERFECT ENZYME ENGINEERING.
J.Biol.Chem., 287, 2012
|
|
3UZW
 
 | |
2CXG
 
 | | CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED TO THE INHIBITOR ACARBOSE | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Strokopytov, B.V, Uitdehaag, J.C.M, Ruiterkamp, R, Dijkstra, B.W. | | Deposit date: | 1998-05-08 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of cyclodextrin glycosyltransferase complexed with acarbose. Implications for the catalytic mechanism of glycosidases.
Biochemistry, 34, 1995
|
|
3UZY
 
 | | Crystal structure of 5beta-reductase (AKR1D1) E120H mutant in complex with NADP+ and 5beta-dihydrotestosterone | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, 5-beta-DIHYDROTESTOSTERONE, CHLORIDE ION, ... | | Authors: | Chen, M, Christianson, D.W, Penning, T.M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | Conversion of Human Steroid 5beta-Reductase (AKR1D1) into 3β-Hydroxysteroid Dehydrogenase by Single Point Mutation E120H: EXAMPLE OF PERFECT ENZYME ENGINEERING.
J.Biol.Chem., 287, 2012
|
|
1VYQ
 
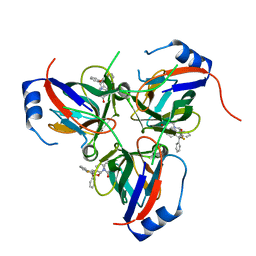 | | Novel inhibitors of Plasmodium Falciparum dUTPase provide a platform for anti-malarial drug design | | Descriptor: | 2,3-DEOXY-3-FLUORO-5-O-TRITYLURIDINE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Whittingham, J.L, Leal, I, Kasinathan, G, Nguyen, C, Bell, E, Jones, A.F, Berry, C, Benito, A, Turkenburg, J.P, Dodson, E.J, Ruiz Perez, L.M, Wilkinson, A.J, Johansson, N.G, Brun, R, Gilbert, I.H, Gonzalez Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dutpase as a Platform for Antimalarial Drug Design: Structural Basis for the Selectivity of a Class of Nucleoside Inhibitors.
Structure, 13, 2005
|
|
3VFW
 
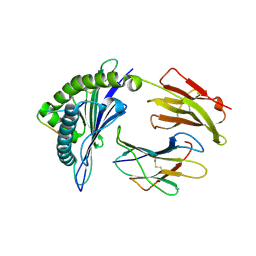 | | crystal structure of HLA B*3508 LPEP-P10Ala, peptide mutant P10-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P10A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VFV
 
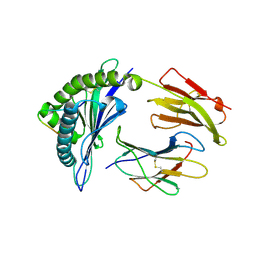 | | crystal structure of HLA B*3508 LPEP-P9Ala, peptide mutant P9-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P9A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
2FO7
 
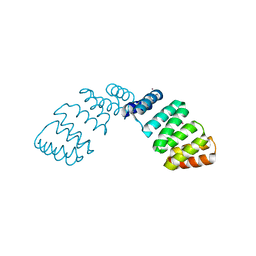 | |
