5KAX
 
 | | The structure of CTR107 protein bound to RHODAMINE 6G | | Descriptor: | CTR107 protein, GLYCEROL, RHODAMINE 6G | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-24 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
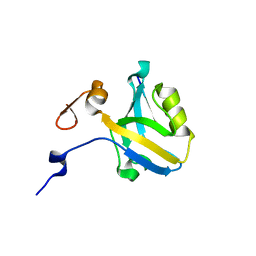
5HFB
 
 | |
3DAY
 
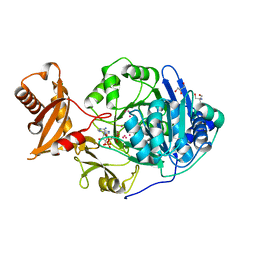 | | Crystal structure of human acyl-CoA synthetase medium-chain family member 2A (L64P mutation) in complex with AMP-CPP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acyl-coenzyme A synthetase ACSM2A, mitochondrial precursor, ... | | Authors: | Pilka, E.S, Kochan, G.T, Yue, W.W, Bhatia, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wikstrom, M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl-coenzyme A synthetase ACSM2A.
J.Mol.Biol., 388, 2009
|
|
5FHT
 
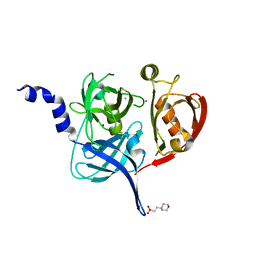 | | HtrA2 protease mutant V226K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Golik, P, Dubin, G, Zurawa-Janicka, D, Lipinska, B, Jarzab, M, Wenta, T, Gieldon, A, Ciarkowski, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Distinct 3D Architecture and Dynamics of the Human HtrA2(Omi) Protease and Its Mutated Variants.
Plos One, 11, 2016
|
|
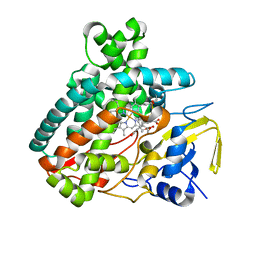
5L1W
 
 | |
1A4A
 
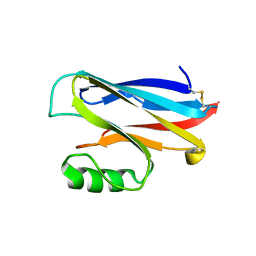 | |
2AUO
 
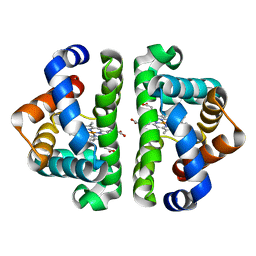 | | Residue F4 plays a key role in modulating the oxygen affinity and cooperatrivity in Scapharca dimeric hemoglobin | | Descriptor: | CARBON MONOXIDE, Globin I, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Bonham, M.A, Gibson, Q.H, Nichols, J.C, Royer Jr, W.E. | | Deposit date: | 2005-08-28 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Residue F4 plays a key role in modulating oxygen affinity and cooperativity in Scapharca dimeric hemoglobin
Biochemistry, 44, 2005
|
|
6CZG
 
 | |
6SPO
 
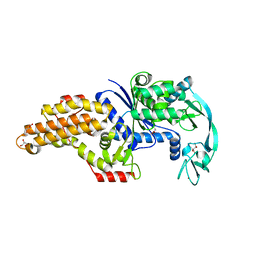 | | Structure of the Escherichia coli methionyl-tRNA synthetase complexed with methionine | | Descriptor: | CITRIC ACID, GLYCEROL, METHIONINE, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
5ONH
 
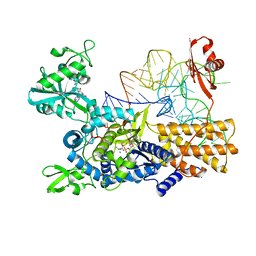 | | Quaternary complex of wild type E. coli leucyl-tRNA synthetase with tRNA(leu), leucyl-adenylate analogue, and post-transfer editing analogue of norvaline in the aminoacylation conformation | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, L-leucyl-tRNA(Leu), ... | | Authors: | Palencia, A, Cusack, S. | | Deposit date: | 2017-08-03 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Kinetic Origin of Substrate Specificity in Post-Transfer Editing by Leucyl-tRNA Synthetase.
J. Mol. Biol., 430, 2018
|
|
5ZH4
 
 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-7 | | Descriptor: | (3R)-6,8-dihydroxy-3-{[(2S,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, CHLORIDE ION, LYSINE, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
5L1V
 
 | |
1A4B
 
 | |
5L1R
 
 | | X-ray Structure of the Substrate-free Cytochrome P450 PntM | | Descriptor: | BICINE, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
1EF2
 
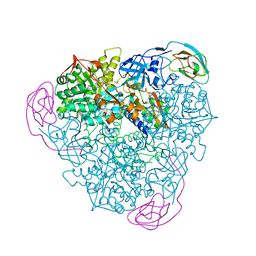 | | CRYSTAL STRUCTURE OF MANGANESE-SUBSTITUTED KLEBSIELLA AEROGENES UREASE | | Descriptor: | MANGANESE (II) ION, UREASE ALPHA SUBUNIT, UREASE BETA SUBUNIT, ... | | Authors: | Yamaguchi, K, Cosper, N.J, Stalhandske, C, Scott, R.A, Pearson, M.A, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 2000-02-05 | | Release date: | 2000-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of metal-substituted Klebsiella aerogenes urease.
J.Biol.Inorg.Chem., 4, 1999
|
|
5L1O
 
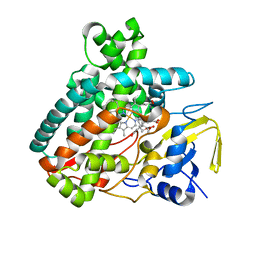 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone F | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone F | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1Q
 
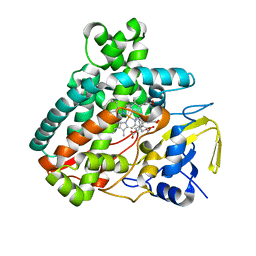 | | X-ray Structure of Cytochrome P450 PntM with Dihydropentalenolactone F | | Descriptor: | Dihydropentalenolactone F, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
1A4C
 
 | | AZURIN MUTANT WITH MET 121 REPLACED BY HIS, PH 3.5 CRYSTAL FORM, DATA COLLECTED AT-180 DEGREES CELSIUS | | Descriptor: | AZURIN, COPPER (II) ION, NITRATE ION, ... | | Authors: | Messerschmidt, A, Prade, L. | | Deposit date: | 1998-01-28 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Rack-induced metal binding vs. flexibility: Met121His azurin crystal structures at different pH.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
5A0Q
 
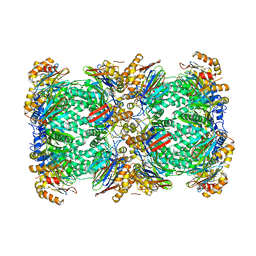 | |
5L1S
 
 | |
1AJB
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE D153G MUTANT OF E. COLI ALKALINE PHOSPHATASE: A MUTANT WITH WEAKER MAGNESIUM BINDING AND INCREASED CATALYTIC ACTIVITY | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Dealwis, C.G, Chen, L, Abad-Zapatero, C. | | Deposit date: | 1995-08-19 | | Release date: | 1995-11-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3-D structure of the D153G mutant of Escherichia coli alkaline phosphatase: an enzyme with weaker magnesium binding and increased catalytic activity.
Protein Eng., 8, 1995
|
|
1FX9
 
 | | CARBOXYLIC ESTER HYDROLASE COMPLEX (DIMERIC PLA2 + MJ33 INHIBITOR + SULPHATE IONS) | | Descriptor: | 1-HEXADECYL-3-TRIFLUOROETHYL-SN-GLYCERO-2-PHOSPHATE METHANE, CALCIUM ION, PHOSPHOLIPASE A2, ... | | Authors: | Pan, Y.H, Epstein, T.M, Jain, M.K, Bahnson, B.J. | | Deposit date: | 2000-09-25 | | Release date: | 2001-09-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Five coplanar anion binding sites on one face of phospholipase A2: relationship to interface binding.
Biochemistry, 40, 2001
|
|
4MW0
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea (Chem 1392) | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
5ZH3
 
 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-6 | | Descriptor: | (3S)-6,8-dihydroxy-3-{[(2R,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, LYSINE, Lysine-tRNA ligase | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
3MSN
 
 | | Crystal structure of Thermolysin in complex with N-methylurea | | Descriptor: | CALCIUM ION, GLYCEROL, N-METHYLUREA, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
