1IA8
 
 | | THE 1.7 A CRYSTAL STRUCTURE OF HUMAN CELL CYCLE CHECKPOINT KINASE CHK1 | | Descriptor: | CHK1 CHECKPOINT KINASE, SULFATE ION | | Authors: | Chen, P, Luo, C, Deng, Y, Ryan, K, Register, J, Margosiak, S, Tempczyk-Russell, A, Nguyen, B, Myers, P, Lundgren, K, Chen Kan, C.-C, O'Connor, P.M. | | Deposit date: | 2001-03-22 | | Release date: | 2001-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation.
Cell(Cambridge,Mass.), 100, 2000
|
|
4OJJ
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P212121) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
1J23
 
 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain | | Descriptor: | ATP-dependent RNA helicase, putative | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
1J25
 
 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain, Mn cocrystal | | Descriptor: | ATP-dependent RNA helicase, putative, MANGANESE (II) ION | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
2YTX
 
 | | Solution structure of the second cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
2YTV
 
 | | Solution structure of the fifth cold-shock domain of the human KIAA0885 protein (unr protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tochio, N, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
4BLV
 
 | | Crystal structure of Escherichia coli 23S rRNA (A2030-N6)- methyltransferase RlmJ in complex with S-adenosylmethionine (AdoMet) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Punekar, A.S, Liljeruhm, J, Shepherd, T.R, Forster, A.C, Selmer, M. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights Into the Molecular Mechanism of Rrna M6A Methyltransferase Rlmj.
Nucleic Acids Res., 41, 2013
|
|
2Z69
 
 | |
3VLF
 
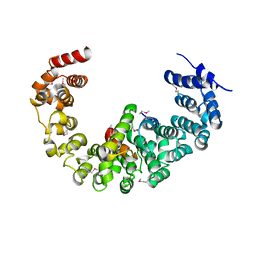 | | Crystal structure of yeast proteasome interacting protein | | Descriptor: | 26S protease regulatory subunit 7 homolog, DNA mismatch repair protein HSM3 | | Authors: | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VZH
 
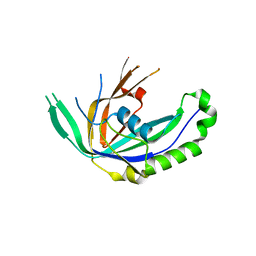 | | Crystal Structure of CRISPR-associated Protein | | Descriptor: | Putative uncharacterized protein | | Authors: | Bae, E, Yoon, K. | | Deposit date: | 2012-10-12 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conservation and Variability in the Structure and Function of the Cas5d Endoribonuclease in the CRISPR-Mediated Microbial Immune System
J.Mol.Biol., 425, 2013
|
|
1AKP
 
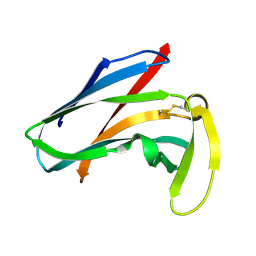 | | SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATION OF APOKEDARCIDIN | | Descriptor: | APOKEDARCIDIN | | Authors: | Constantine, K.L, Colson, K.L, Wittekind, M, Friedrichs, M.S, Zein, N, Tuttle, J, Langley, D.R, Leet, J.E, Schroeder, D.R, Lam, K.S, Farmer II, B.T, Metzler, W.J, Bruccoleri, R.E, Mueller, L. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Sequential 1H, 13C, and 15N NMR assignments and solution conformation of apokedarcidin.
Biochemistry, 33, 1994
|
|
4BLU
 
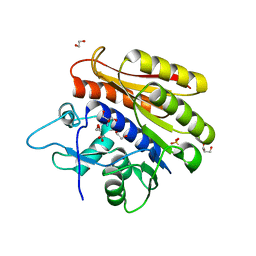 | | Crystal structure of Escherichia coli 23S rRNA (A2030-N6)- methyltransferase RlmJ | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Punekar, A.S, Liljeruhm, J, Shepherd, T.R, Forster, A.C, Selmer, M. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Insights Into the Molecular Mechanism of Rrna M6A Methyltransferase Rlmj.
Nucleic Acids Res., 41, 2013
|
|
4BLW
 
 | | Crystal structure of Escherichia coli 23S rRNA (A2030-N6)- methyltransferase RlmJ in complex with S-adenosylhomocysteine (AdoHcy) and Adenosine monophosphate (AMP) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Punekar, A.S, Liljeruhm, J, Shepherd, T.R, Forster, A.C, Selmer, M. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Insights Into the Molecular Mechanism of Rrna M6A Methyltransferase Rlmj.
Nucleic Acids Res., 41, 2013
|
|
3VZI
 
 | | Crystal Structure of CRISPR-associated Protein | | Descriptor: | Crispr-associated protein Cas5, dvulg subtype | | Authors: | Bae, E, Koo, Y. | | Deposit date: | 2012-10-12 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Conservation and Variability in the Structure and Function of the Cas5d Endoribonuclease in the CRISPR-Mediated Microbial Immune System
J.Mol.Biol., 425, 2013
|
|
2EKE
 
 | |
1OFT
 
 | |
1P1A
 
 | | NMR structure of ubiquitin-like domain of hHR23B | | Descriptor: | UV excision repair protein RAD23 homolog B | | Authors: | Ryu, K.S, Lee, K.J, Bae, S.H, Kim, B.K, Kim, K.A, Choi, B.S. | | Deposit date: | 2003-04-11 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a
J.Biol.Chem., 278, 2003
|
|
1NLN
 
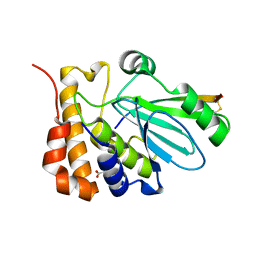 | | CRYSTAL STRUCTURE OF HUMAN ADENOVIRUS 2 PROTEINASE WITH ITS 11 AMINO ACID COFACTOR AT 1.6 ANGSTROM RESOLUTION | | Descriptor: | ACETIC ACID, Adenain, PVIC | | Authors: | McGrath, W.J, Ding, J, Sweet, R.M, Mangel, W.F. | | Deposit date: | 2003-01-07 | | Release date: | 2003-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic structure at 1.6-A resolution of the human adenovirus proteinase in a covalent complex with its 11-amino-acid peptide cofactor: insights on a new fold
Biochim.Biophys.Acta, 1648, 2003
|
|
3J4B
 
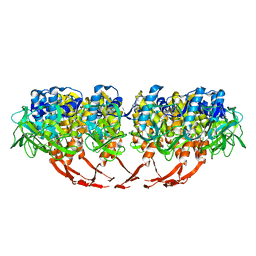 | | Structure of T7 gatekeeper protein (gp11) | | Descriptor: | Tail tubular protein A | | Authors: | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
4ESF
 
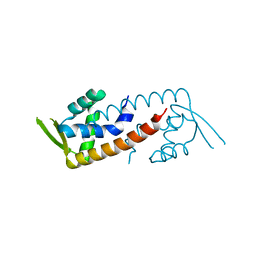 | |
2YGV
 
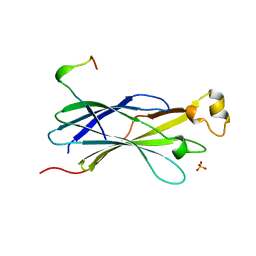 | | Conserved N-terminal domain of the yeast Histone Chaperone Asf1 in complex with the C-terminal fragment of Rad53 | | Descriptor: | GLYCEROL, HISTONE CHAPERONE ASF1, SERINE/THREONINE-PROTEIN KINASE RAD53, ... | | Authors: | Jiao, Y, Seeger, K, Murciano, B, Ledu, M.H, Charbonnier, J.B, Legrand, P, Lautrette, A, Gaubert, A, Mousson, F, Guerois, R, Mann, C, Ochsenbein, F. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Surprising Complexity of the Asf1 Histone Chaperone-Rad53 Kinase Interaction
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1YDX
 
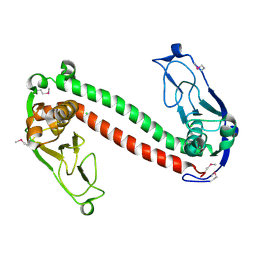 | | Crystal structure of Type-I restriction-modification system S subunit from M. genitalium | | Descriptor: | CHLORIDE ION, type I restriction enzyme specificity protein MG438 | | Authors: | Machado, B, Quijada, O, Pinol, J, Fita, I, Querol, E, Carpena, X. | | Deposit date: | 2004-12-27 | | Release date: | 2005-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Putative Type I Restriction-Modification S Subunit from Mycoplasma genitalium
J.Mol.Biol., 351, 2005
|
|
1L1L
 
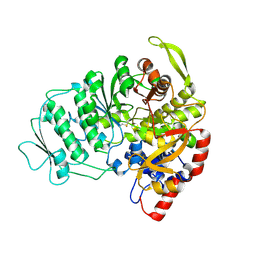 | | CRYSTAL STRUCTURE OF B-12 DEPENDENT (CLASS II) RIBONUCLEOTIDE REDUCTASE | | Descriptor: | RIBONUCLEOSIDE TRIPHOSPHATE REDUCTASE | | Authors: | Sintchak, M.D, Arjara, G, Kellogg, B.A, Stubbe, J, Drennan, C.L. | | Deposit date: | 2002-02-18 | | Release date: | 2002-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of class II ribonucleotide reductase reveals how an allosterically regulated monomer mimics a dimer.
Nat.Struct.Biol., 9, 2002
|
|
2BRJ
 
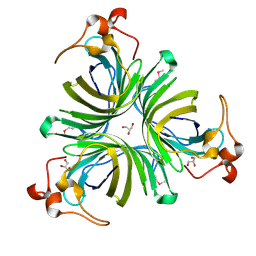 | | X-ray structure of the Allene Oxide Cyclase from Arabidopsis thaliana | | Descriptor: | ARABIDOPSIS THALIANA GENOMIC DNA, CHROMOSOME 3,, GLYCEROL | | Authors: | Hofmann, E, Zerbe, P, Schaller, F. | | Deposit date: | 2005-05-06 | | Release date: | 2006-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of Arabidopsis Thaliana Allene Oxide Cyclase: Insights Into the Oxylipin Cyclization Reaction.
Plant Cell, 18, 2006
|
|
4BV6
 
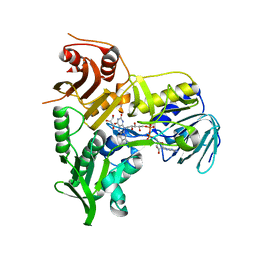 | | Refined crystal structure of the human Apoptosis inducing factor | | Descriptor: | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Hermoso, J.A, Ferreira, P, Villanueva, R, Medina, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Coenzyme Mediated Monomer-Dimer Transition of the Pro-Apoptotic Apoptosis Inducing Factor.
Biochemistry, 53, 2014
|
|
