1QUA
 
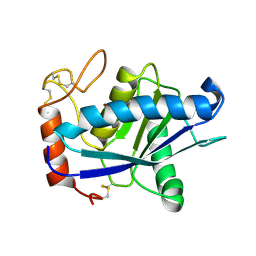 | | CRYSTAL STRUCTURE OF ACUTOLYSIN-C, A HEMORRHAGIC TOXIN FROM THE SNAKE VENOM OF AGKISTRODON ACUTUS, AT 2.2 A RESOLUTION | | Descriptor: | ACUTOLYSIN-C, ZINC ION | | Authors: | Niu, L, Teng, M, Zhu, X. | | Deposit date: | 1999-06-30 | | Release date: | 2000-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of acutolysin-C, a haemorrhagic toxin from the venom of Agkistrodon acutus, providing further evidence for the mechanism of the pH-dependent proteolytic reaction of zinc metalloproteinases.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1QTJ
 
 | |
5KLT
 
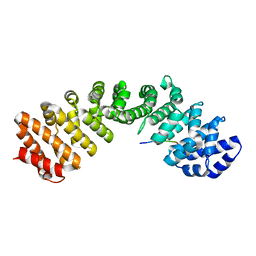 | | Prototypical P4[M]cNLS | | Descriptor: | Importin subunit alpha-1, Prototypical P4[M]cNLS | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2016-06-25 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Contribution of the residue at position 4 within classical nuclear localization signals to modulating interaction with importins and nuclear targeting.
Biochim Biophys Acta Mol Cell Res, 1865, 2018
|
|
1QXH
 
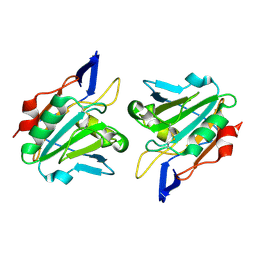 | |
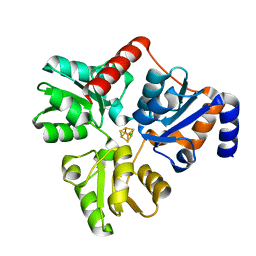
5KTM
 
 | |
5KTS
 
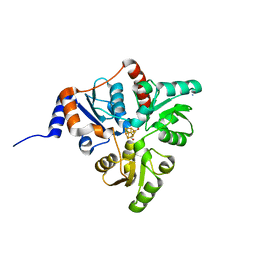 | | Crystal structure of Pyrococcus horikoshii quinolinate synthase (NadA) with bound citraconate and Fe4S4 cluster | | Descriptor: | (~{Z})-2-methylbut-2-enedioic acid, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Ealick, S.E. | | Deposit date: | 2016-07-12 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structures of the Iron-Sulfur Cluster-Dependent Quinolinate Synthase in Complex with Dihydroxyacetone Phosphate, Iminoaspartate Analogues, and Quinolinate.
Biochemistry, 55, 2016
|
|
1QOR
 
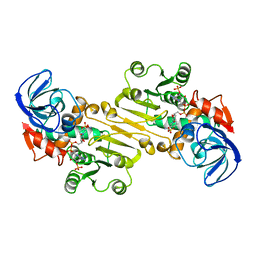 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI QUINONE OXIDOREDUCTASE COMPLEXED WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, QUINONE OXIDOREDUCTASE, SULFATE ION | | Authors: | Thorn, J.M, Barton, J.D, Dixon, N.E, Ollis, D.L, Edwards, K.J. | | Deposit date: | 1995-02-14 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Escherichia coli QOR quinone oxidoreductase complexed with NADPH.
J.Mol.Biol., 249, 1995
|
|
3TWQ
 
 | |
3TWW
 
 | |
1QP1
 
 | | KAPPA VARIABLE LIGHT CHAIN | | Descriptor: | BENCE-JONES KAPPA I ANTIBODY BRE (LIGHT CHAIN) | | Authors: | Steinrauf, L.K. | | Deposit date: | 1999-05-30 | | Release date: | 1999-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular structure of the amyloid-forming protein kappa I Bre.
J.Biochem.(Tokyo), 125, 1999
|
|
1QNF
 
 | | STRUCTURE OF PHOTOLYASE | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, FLAVIN-ADENINE DINUCLEOTIDE, PHOTOLYASE | | Authors: | Miki, K, Kitadokoro, K. | | Deposit date: | 1997-07-04 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of DNA photolyase from Anacystis nidulans
Nat.Struct.Biol., 4, 1997
|
|
1QA4
 
 | |
1J9T
 
 | | Crystal structure of nitrite soaked reduced H255N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
1QCE
 
 | |
3TKG
 
 | | crystal structure of HIV model protease precursor/saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | Deposit date: | 2011-08-26 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
2P1L
 
 | |
1NNS
 
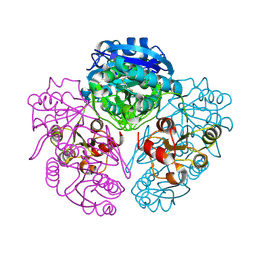 | | L-asparaginase of E. coli in C2 space group and 1.95 A resolution | | Descriptor: | ASPARTIC ACID, L-asparaginase II | | Authors: | Sanches, M, Barbosa, J.A.R.G, de Oliveira, R.T, Neto, J.A.A, Polikarpov, I. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural comparison of Escherichia coli L-asparaginase in two monoclinic space groups.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6AN8
 
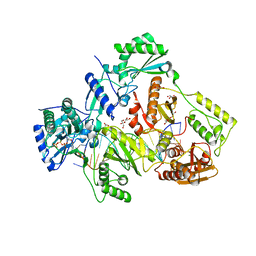 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 8.0 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA PRIMER (5'- D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*GP)-3'), DNA TEMPLATE (5'- D(*AP*TP*GP*AP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
2CAU
 
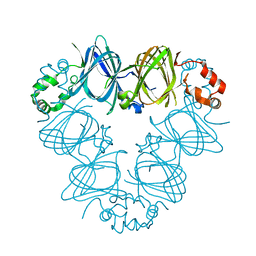 | | CANAVALIN FROM JACK BEAN | | Descriptor: | PROTEIN (CANAVALIN) | | Authors: | Ko, T.-P, Day, J, Macpherson, A. | | Deposit date: | 1998-11-20 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2.0 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3RTN
 
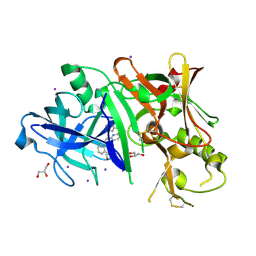 | |
2CAV
 
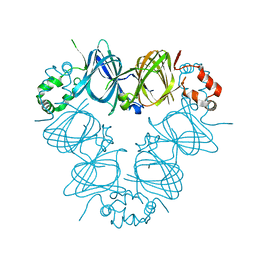 | | CANAVALIN FROM JACK BEAN | | Descriptor: | PROTEIN (CANAVALIN) | | Authors: | Ko, T.-P, Day, J, Macpherson, A. | | Deposit date: | 1998-11-20 | | Release date: | 1998-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2.0 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
8Q6J
 
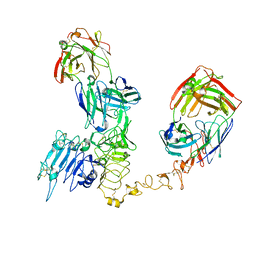 | | Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, Pertuzumab Fab light chain, ... | | Authors: | Ruedas, R, Vuillemot, R, Tubiana, T, Winter, J.M, Pieri, L, Arteni, A.A, Samson, C, Jonic, J, Mathieu, M, Bressanelli, S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and conformational variability of the HER2-trastuzumab-pertuzumab complex.
J.Struct.Biol., 216, 2024
|
|
1P58
 
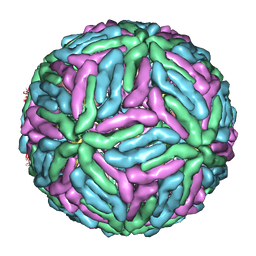 | | Complex Organization of Dengue Virus Membrane Proteins as Revealed by 9.5 Angstrom Cryo-EM reconstruction | | Descriptor: | Envelope protein M, Major envelope protein E | | Authors: | Zhang, W, Chipman, P.R, Corver, J, Johnson, P.R, Zhang, Y, Mukhopadhyay, S, Baker, T.S, Strauss, J.H, Rossmann, M.G, Kuhn, R.J. | | Deposit date: | 2003-04-25 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Visualization of membrane protein domains by cryo-electron microscopy of dengue virus
Nat.Struct.Biol., 10, 2003
|
|
2CDV
 
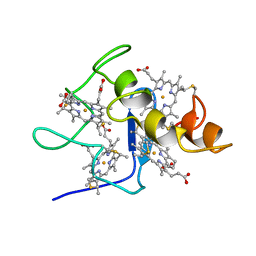 | | REFINED STRUCTURE OF CYTOCHROME C3 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Higuchi, Y, Kusunoki, M, Matsuura, Y, Yasuoka, N, Kakudo, M. | | Deposit date: | 1983-11-15 | | Release date: | 1984-02-02 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined structure of cytochrome c3 at 1.8 A resolution
J.Mol.Biol., 172, 1984
|
|
4DYX
 
 | |
