1RMQ
 
 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase with osmiate mimicking the catalytic intermediate | | Descriptor: | COBALT (II) ION, Class B acid phosphatase, OSMIUM ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights in the catalytic mechanism of AphA from Escherichia coli
To be Published
|
|
1RMR
 
 | | Crystal Structure of Schistatin, a Disintegrin Homodimer from saw-scaled Viper (Echis carinatus) at 2.5 A resolution | | Descriptor: | Disintegrin schistatin | | Authors: | Bilgrami, S, Tomar, S, Yadav, S, Kaur, P, Kumar, J, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of schistatin, a disintegrin homodimer from saw-scaled viper (Echis carinatus) at 2.5 A resolution
J.Mol.Biol., 341, 2004
|
|
1RMS
 
 | | CRYSTAL STRUCTURES OF RIBONUCLEASE MS COMPLEXED WITH 3'-GUANYLIC ACID A GP*C ANALOGUE, 2'-DEOXY-2'-FLUOROGUANYLYL-3',5'-CYTIDINE | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE MS | | Authors: | Nonaka, T, Mitsui, Y, Nakamura, K.T. | | Deposit date: | 1991-12-02 | | Release date: | 1992-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ribonuclease Ms (as a ribonuclease T1 homologue) complexed with a guanylyl-3',5'-cytidine analogue.
Biochemistry, 32, 1993
|
|
1RMT
 
 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase complexed with adenosine. | | Descriptor: | ADENOSINE, Class B acid phosphatase, MAGNESIUM ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights in the catalytic mechanism of AphA from Escherichia coli
To be Published
|
|
1RMU
 
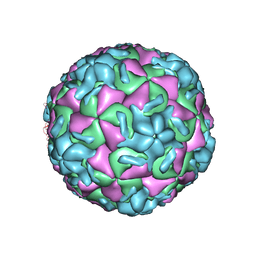 | | THREE-DIMENSIONAL STRUCTURES OF DRUG-RESISTANT MUTANTS OF HUMAN RHINOVIRUS 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Krishnaswamy, S, Kremer, M.J, Oliveira, M.A, Rossmann, M.G, Heinz, B.A, Rueckert, R.R, Dutko, F.J, Mckinlay, M.A. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of drug-resistant mutants of human rhinovirus 14.
J.Mol.Biol., 207, 1989
|
|
1RMV
 
 | | RIBGRASS MOSAIC VIRUS, FIBER DIFFRACTION | | Descriptor: | RIBGRASS MOSAIC VIRUS COAT PROTEIN, RIBGRASS MOSAIC VIRUS RNA | | Authors: | Wang, H, Stubbs, G. | | Deposit date: | 1997-02-11 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | FIBER DIFFRACTION (2.9 Å) | | Cite: | Molecular dynamics in refinement against fiber diffraction data.
Acta Crystallogr.,Sect.A, 49, 1993
|
|
1RMX
 
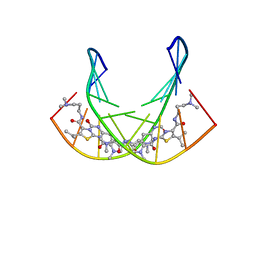 | | A SHORT LEXITROPSIN THAT RECOGNIZES THE DNA MINOR GROOVE AT 5'-ACTAGT-3': UNDERSTANDING THE ROLE OF ISOPROPYL-THIAZOLE | | Descriptor: | 5'-D(*CP*GP*AP*CP*TP*AP*GP*TP*CP*G)-3', N-[3-(DIMETHYLAMINO)PROPYL]-2-({[4-({[4-(FORMYLAMINO)-1-METHYL-1H-PYRROL-2-YL]CARBONYL}AMINO)-1-METHYL-1H-PYRROL-2-YL]CARBONYL}AMINO)-5-ISOPROPYL-1,3-THIAZOLE-4-CARBOXAMIDE | | Authors: | Anthony, N.G, Johnston, B.F, Khalaf, A.I, Mackay, S.P, Parkinson, J.A, Suckling, C.J, Waigh, R.D. | | Deposit date: | 2003-11-28 | | Release date: | 2004-08-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Short Lexitropsin that Recognizes the DNA Minor Groove at 5'-ACTAGT-3': Understanding the Role of Isopropyl-thiazole.
J.Am.Chem.Soc., 126, 2004
|
|
1RMY
 
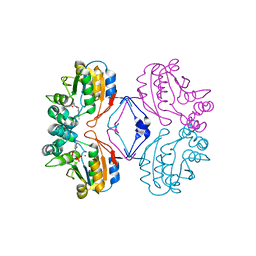 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase ternary complex with deoxycytosine and phosphate bound to the catalytic metal | | Descriptor: | 2'-DEOXYCYTIDINE, Class B acid phosphatase, MAGNESIUM ION, ... | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights in the catalytic mechanism of AphA from Escherichia coli
To be Published
|
|
1RMZ
 
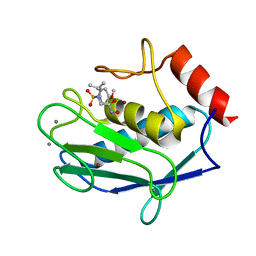 | | Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor NNGH at 1.3 A resolution | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mangani, S, Terni, B. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Conformational variability of matrix metalloproteinases: beyond a single 3D structure.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1RN1
 
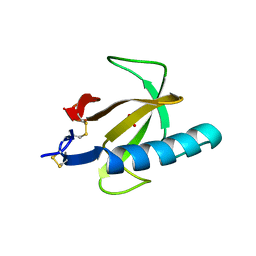 | | THREE-DIMENSIONAL STRUCTURE OF GLN 25-RIBONUCLEASE T1 AT 1.84 ANGSTROMS RESOLUTION: STRUCTURAL VARIATIONS AT THE BASE RECOGNITION AND CATALYTIC SITES | | Descriptor: | RIBONUCLEASE T1 ISOZYME, SULFATE ION | | Authors: | Arni, R.K, Pal, G.P, Ravichandran, K.G, Tulinsky, A, Walz Junior, F.G, Metcalf, P. | | Deposit date: | 1991-11-22 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Three-dimensional structure of Gln25-ribonuclease T1 at 1.84-A resolution: structural variations at the base recognition and catalytic sites.
Biochemistry, 31, 1992
|
|
1RN4
 
 | |
1RN7
 
 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-11-30 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
1RN8
 
 | | Crystal structure of dUTPase complexed with substrate analogue imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
1RN9
 
 | | A SHORT LEXITROPSIN THAT RECOGNIZES THE DNA MINOR GROOVE AT 5'-ACTAGT-3': UNDERSTANDING THE ROLE OF ISOPROPYL-THIAZOLE | | Descriptor: | 5'-D(*CP*GP*AP*CP*TP*AP*GP*TP*CP*G)-3' | | Authors: | Anthony, N.G, Johnston, B.F, Khalaf, A.I, Mackay, S.P, Parkinson, J.A, Suckling, C.J, Waigh, R.D. | | Deposit date: | 2003-12-01 | | Release date: | 2004-08-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Short Lexitropsin that Recognizes the DNA Minor Groove at 5'-ACTAGT-3': Understanding the Role of Isopropyl-thiazole.
J.Am.Chem.Soc., 126, 2004
|
|
1RNA
 
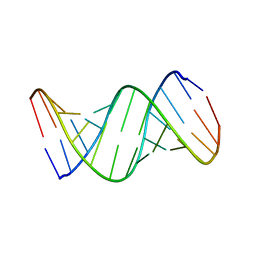 | | CRYSTALLOGRAPHIC STRUCTURE OF AN RNA HELIX: [U(U-A)6A]2 | | Descriptor: | RNA (5'-R(*UP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*A)-3') | | Authors: | Dock-Bregeon, A.C, Chevrier, B, Podjarny, A, Johnson, J, De Bear, J.S, Gough, G.R, Gilham, P.T, Moras, D. | | Deposit date: | 1990-02-01 | | Release date: | 1991-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic structure of an RNA helix: [U(UA)6A]2.
J.Mol.Biol., 209, 1989
|
|
1RNB
 
 | |
1RNC
 
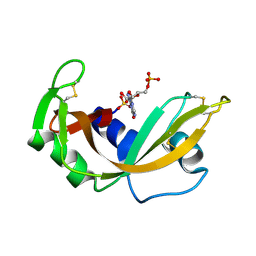 | | NEWLY OBSERVED BINDING MODE IN PANCREATIC RIBONUCLEASE | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE A, SULFATE ION | | Authors: | Aguilar, C.F, Thomas, P.J, Mills, A, Moss, D.S, Palmer, R.A. | | Deposit date: | 1991-10-21 | | Release date: | 1994-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Newly observed binding mode in pancreatic ribonuclease.
J.Mol.Biol., 224, 1992
|
|
1RND
 
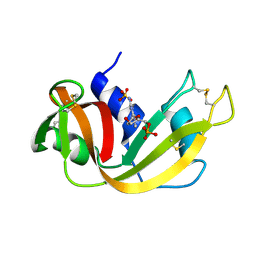 | | NEWLY OBSERVED BINDING MODE IN PANCREATIC RIBONUCLEASE | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE A, SULFATE ION | | Authors: | Aguilar, C.F, Thomas, P.J, Mills, A, Moss, D.S, Palmer, R.A. | | Deposit date: | 1991-10-21 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Newly observed binding mode in pancreatic ribonuclease.
J.Mol.Biol., 224, 1992
|
|
1RNE
 
 | |
1RNF
 
 | | X-RAY CRYSTAL STRUCTURE OF UNLIGANDED HUMAN RIBONUCLEASE 4 | | Descriptor: | PROTEIN (RIBONUCLEASE 4) | | Authors: | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Rueth, F.X, Coll, M. | | Deposit date: | 1998-10-29 | | Release date: | 1999-10-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
1RNG
 
 | |
1RNH
 
 | | STRUCTURE OF RIBONUCLEASE H PHASED AT 2 ANGSTROMS RESOLUTION BY MAD ANALYSIS OF THE SELENOMETHIONYL PROTEIN | | Descriptor: | RIBONUCLEASE HI, SULFATE ION | | Authors: | Yang, W, Hendrickson, W.A, Crouch, R.J, Satow, Y. | | Deposit date: | 1990-07-11 | | Release date: | 1991-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein.
Science, 249, 1990
|
|
1RNI
 
 | | Bifunctional DNA primase/polymerase domain of ORF904 from the archaeal plasmid pRN1 | | Descriptor: | ORF904, ZINC ION | | Authors: | Lipps, G, Weinzierl, A.O, von Scheven, G, Buchen, C, Cramer, P. | | Deposit date: | 2003-12-01 | | Release date: | 2004-01-27 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a bifunctional DNA primase-polymerase
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RNJ
 
 | | Crystal structure of inactive mutant dUTPase complexed with substrate analogue imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
1RNK
 
 | |
