4FNY
 
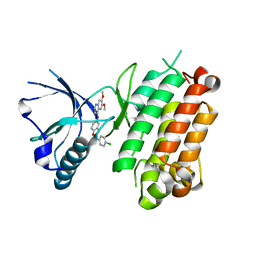 | | Crystal structure of the R1275Q anaplastic lymphoma kinase catalytic domain in complex with a benzoxazole inhibitor | | Descriptor: | ALK tyrosine kinase receptor, N-(4-chlorophenyl)-5-[(6,7-dimethoxyquinolin-4-yl)oxy]-1,3-benzoxazol-2-amine | | Authors: | Whittington, D.A, Epstein, L.F, Chen, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The R1275Q Neuroblastoma Mutant and Certain ATP-competitive Inhibitors Stabilize Alternative Activation Loop Conformations of Anaplastic Lymphoma Kinase.
J.Biol.Chem., 287, 2012
|
|
2BGY
 
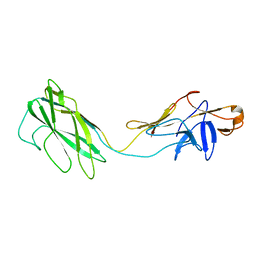 | | Fit of the x-ray structure of the baterial flagellar hook fragment flge31 into an EM map from the hook of Caulobacter crescentus. | | Descriptor: | FLAGELLAR HOOK PROTEIN FLGE | | Authors: | Shaikh, T.R, Thomas, D.R, Chen, J.Z, Samatey, F.A, Matsunami, H, Imada, K, Namba, K, DeRosier, D.J. | | Deposit date: | 2005-01-06 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | A Partial Atomic Structure for the Flagellar Hook of Salmonella Typhimurium.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5DG0
 
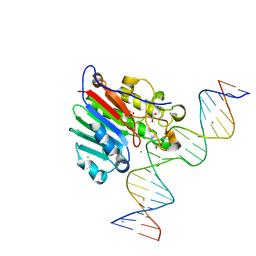 | |
5DGM
 
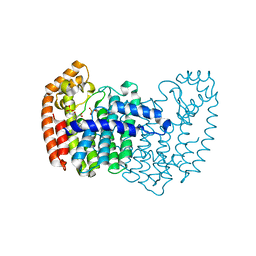 | |
2C1J
 
 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | Authors: | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | Deposit date: | 2005-09-15 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
5DEG
 
 | | Crystal structure of B*27:06 bound to the pVIPR peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Loll, B, Fabian, H, Hee, C.S, Huser, H, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Increased Conformational Flexibility of HLA-B*27 Subtypes Associated With Ankylosing Spondylitis.
Arthritis Rheumatol, 68, 2016
|
|
4FDG
 
 | | Crystal Structure of an Archaeal MCM Filament | | Descriptor: | Minichromosome maintenance protein MCM, ZINC ION | | Authors: | Slaymaker, I.M, Fu, Y, Brewster, A.B, Chen, X.S. | | Deposit date: | 2012-05-28 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Mini-chromosome maintenance complexes form a filament to remodel DNA structure and topology.
Nucleic Acids Res., 41, 2013
|
|
4FFM
 
 | | PylC in complex with L-lysine-Ne-D-ornithine (cocrystallized with L-lysine-Ne-D-ornithine) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Quitterer, F, List, A, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of PylC at 1.5A resolution.
J.Mol.Biol., 424, 2012
|
|
2BWO
 
 | | 5-Aminolevulinate Synthase from Rhodobacter capsulatus in complex with succinyl-CoA | | Descriptor: | 5-AMINOLEVULINATE SYNTHASE, PYRIDOXAL-5'-PHOSPHATE, SUCCINYL-COENZYME A | | Authors: | Astner, I, Schulze, J.O, van den Heuvel, J.J, Jahn, D, Schubert, W.-D, Heinz, D.W. | | Deposit date: | 2005-07-15 | | Release date: | 2005-09-27 | | Last modified: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of 5-Aminolevulinate Synthase, the First Enzyme of Heme Biosynthesis, and its Link to Xlsa in Humans.
Embo J., 24, 2005
|
|
4FFR
 
 | | SeMet-labeled PylC (remote) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PylC | | Authors: | Quitterer, F, List, A, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of PylC at 1.5A resolution.
J.Mol.Biol., 424, 2012
|
|
5DFI
 
 | | Human APE1 phosphorothioate substrate complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(OMC)P*(48Z)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2BDF
 
 | | Src kinase in complex with inhibitor AP23451 | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, {[(4-{[2-(4-AMINOCYCLOHEXYL)-9-ETHYL-9H-PURIN-6-YL]AMINO}PHENYL)(HYDROXY)PHOSPHORYL]METHYL}PHOSPHONIC ACID | | Authors: | Dalgarno, D, Stehle, T, Schelling, P, Sawyer, T, Narula, S. | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of Src tyrosine kinase inhibition with a new class of potent and selective trisubstituted purine-based compounds.
Chem.Biol.Drug Des., 67, 2006
|
|
5DIY
 
 | | Thermobaculum terrenum O-GlcNAc hydrolase mutant - D120N | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hyaluronidase, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Ostrowski, A, Gundogdu, M, Ferenbach, A.T, Lebedev, A, van Aalten, D.M.F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum.
J.Biol.Chem., 290, 2015
|
|
4FID
 
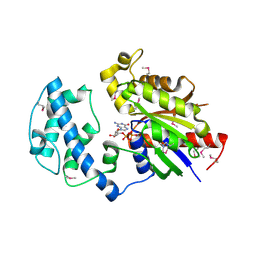 | | Crystal structure of a heterotrimeric G-Protein subunit from entamoeba histolytica, EHG-ALPHA-1 | | Descriptor: | G protein alpha subunit, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Bosch, D.E, Kimple, A.J, Muller, R.E, Gigure, P.M, Willard, F.S, Machius, M, Temple, B.R, Siderovski, D.P. | | Deposit date: | 2012-06-08 | | Release date: | 2012-11-28 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Heterotrimeric G-protein Signaling Is Critical to Pathogenic Processes in Entamoeba histolytica.
Plos Pathog., 8, 2012
|
|
4FL5
 
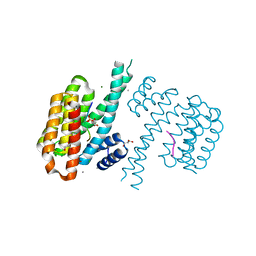 | |
5DIR
 
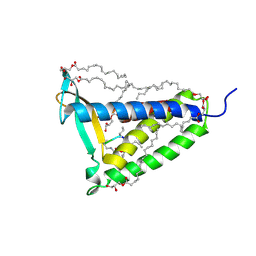 | | membrane protein at 2.8 Angstroms | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Globomycin, Lipoprotein signal peptidase | | Authors: | Vogeley, L, El Arnaout, T, Bailey, J, Boland, C, Caffrey, M. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin.
Science, 351, 2016
|
|
2C0S
 
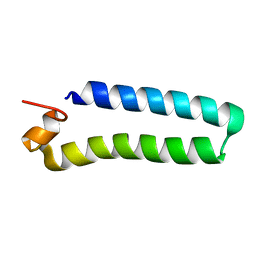 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-09-07 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
5DQU
 
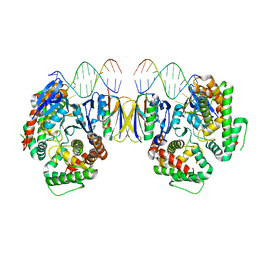 | | Crystal Structure of Cas-DNA-10 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(*GP*AP*GP*TP*CP*GP*AP*TP*GP*CP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
8FG2
 
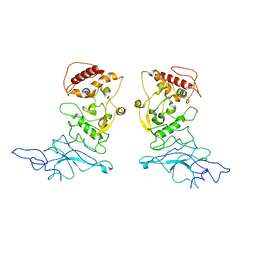 | | SARS-CoV-2 Nucleocapsid dimer structure determined from COVID-19 patients | | Descriptor: | Nucleoprotein | | Authors: | Casasanta, M, Jonaid, G.M, Kaylor, L, Luqiu, W, DiCecco, L, Solares, M, Berry, S, Kelly, D.F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural Insights of the SARS-CoV-2 Nucleocapsid Protein: Implications for the Inner-workings of Rapid Antigen Tests.
Microsc Microanal, 29, 2023
|
|
4FGI
 
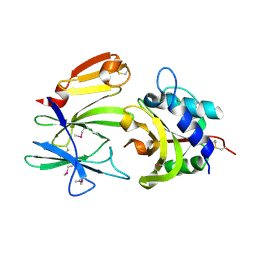 | | Structure of the effector - immunity system Tse1 / Tsi1 from Pseudomonas aeruginosa | | Descriptor: | Tse1, Tsi1 | | Authors: | Benz, J, Sendlmeier, C, Barends, T.R.M, Meinhart, A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-06-20 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tse1/Tsi1 from Pseudomonas aeruginosa.
Plos One, 7, 2012
|
|
2BQX
 
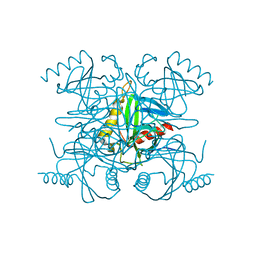 | |
2BHI
 
 | | Crystal structure of Taiwan cobra cardiotoxin A3 complexed with sulfogalactoceramide | | Descriptor: | CYTOTOXIN 3, HEXAETHYLENE GLYCOL MONODECYL ETHER, SULFOGALACTOCERAMIDE | | Authors: | Wang, C.-H, Liu, J.-H, Wu, P.-L, Lee, S.-C, Hsiao, C.-D, Wu, W.-G. | | Deposit date: | 2005-01-12 | | Release date: | 2005-11-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Glycosphingolipid-Facilitated Membrane Insertion and Internalization of Cobra Cardiotoxin: The Sulfatide/Cardiotoxin Complex Structure in a Membrane-Like Environment Suggests a Lipid-Dependent Cell-Penetrating Mechanism for Membrane Binding Polypeptides.
J.Biol.Chem., 281, 2006
|
|
7T21
 
 | | E. coli DnaB bound to ssDNA and ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Oakley, A.J, Xu, Z.Q. | | Deposit date: | 2021-12-02 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Sequence of conformation changes of DnaB helicase during DNA unwinding and priming in Escherichia coli
To Be Published
|
|
5DRO
 
 | | Structure of the Aquifex aeolicus LpxC/LPC-011 Complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
2C4W
 
 | |
