7D2M
 
 | |
4ZLX
 
 | |
6LW3
 
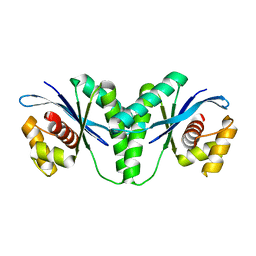 | | Crystal structure of RuvC from Pseudomonas aeruginosa | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC | | Authors: | Hu, Y, He, Y, Lin, Z. | | Deposit date: | 2020-02-07 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Biochemical and structural characterization of the Holliday junction resolvase RuvC from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
3PXE
 
 | |
3NK3
 
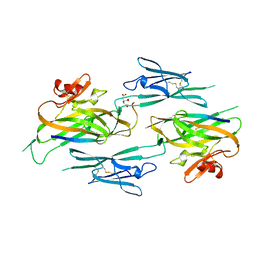 | | Crystal structure of full-length sperm receptor ZP3 at 2.6 A resolution | | Descriptor: | CITRATE ANION, Zona pellucida 3, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Monne, M, Jovine, L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into Egg Coat Assembly and Egg-Sperm Interaction from the X-Ray Structure of Full-Length ZP3.
Cell(Cambridge,Mass.), 143, 2010
|
|
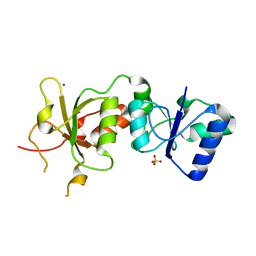
3PXB
 
 | |
3PXC
 
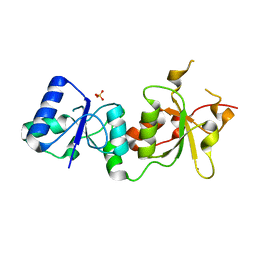 | |
2ADW
 
 | |
2B5N
 
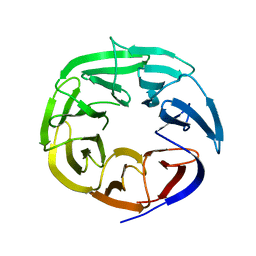 | | Crystal Structure of the DDB1 BPB Domain | | Descriptor: | ISOPROPYL ALCOHOL, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
2AKY
 
 | |
5OFA
 
 | | Crystal structure of human MORC2 (residues 1-603) with spinal muscular atrophy mutation T424R | | Descriptor: | MAGNESIUM ION, MORC family CW-type zinc finger protein 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Douse, C.H, Liu, Y, Modis, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Neuropathic MORC2 mutations perturb GHKL ATPase dimerization dynamics and epigenetic silencing by multiple structural mechanisms.
Nat Commun, 9, 2018
|
|
6JVF
 
 | | Crystal structure of human apo MTH1 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Peng, C, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVL
 
 | | Crystal structure of human MTH1 in complex with compound MI1014 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-5-ethyl-6-(4-methylpiperazin-1-yl)pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6QNZ
 
 | | Crystal structure of the site-specific DNA nickase N.BspD6I E418A Mutant | | Descriptor: | GLYCEROL, Heterodimeric restriction endonuclease R.BspD6I large subunit, PHOSPHATE ION | | Authors: | Artyukh, R.I, Kachalova, G.S, Yunusova, A.K, Gabdulkhakov, A.G, Fatkhullin, B.F, Atanasov, B.P, Perevyazova, T.A, Popov, A.N, Zheleznaya, L.A. | | Deposit date: | 2019-02-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The key role of E418 carboxyl group in the formation of Nt.BspD6I nickase active site: Structural and functional properties of Nt.BspD6I E418A mutant.
J.Struct.Biol., 210, 2020
|
|
6QX7
 
 | |
2J04
 
 | | The tau60-tau91 subcomplex of yeast transcription factor IIIC | | Descriptor: | HYPOTHETICAL PROTEIN YPL007C, YDR362CP | | Authors: | Mylona, A, Fernandez-Tornero, C, Legrand, P, Muller, C.W. | | Deposit date: | 2006-07-31 | | Release date: | 2006-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Tau60/Deltatau91 Subcomplex of Yeast Transcription Factor Iiic: Insights Into Preinitiation Complex Assembly
Mol.Cell, 24, 2006
|
|
4Z2W
 
 | | Factor Inhibiting HIF in Complex with Fe, and Alpha-Ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Taabazuing, C.Y, Garman, S.C, Knapp, M.J. | | Deposit date: | 2015-03-30 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Promotes Productive Gas Binding in the alpha-Ketoglutarate-Dependent Oxygenase FIH.
Biochemistry, 55, 2016
|
|
6QYM
 
 | |
5OFB
 
 | | Crystal structure of human MORC2 (residues 1-603) with spinal muscular atrophy mutation S87L | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MORC family CW-type zinc finger protein 2, ... | | Authors: | Douse, C.H, Liu, Y, Modis, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Neuropathic MORC2 mutations perturb GHKL ATPase dimerization dynamics and epigenetic silencing by multiple structural mechanisms.
Nat Commun, 9, 2018
|
|
4M3I
 
 | | X-ray crystal structure of the ruthenium complex [Ru(TAP)2(dppz-{Me2})]2+ bound to d(CCGGTACCGG) | | Descriptor: | (11,12-dimethyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium(2+), BARIUM ION, Synthetic DNA CCGGTACCGG | | Authors: | Niyazi, H, Teixeira, S, Mitchell, E, Forsyth, T, Cardin, C. | | Deposit date: | 2013-08-06 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of the ruthenium complex [Ru(TAP)2(dppz-{Me2})]2+ bound to d(CCGGTACCGG)
To be Published
|
|
4HNJ
 
 | |
4MMJ
 
 | | crystal structure of YafQ from E.coli strain BL21(DE3) | | Descriptor: | Addiction module toxin, RelE/StbE family, SULFATE ION | | Authors: | Liang, Y, Gao, Z, Liu, Q, Dong, Y. | | Deposit date: | 2013-09-09 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
8FMF
 
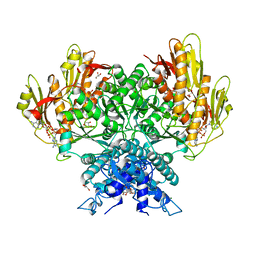 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-diAMP ligand (1 tetramer in the AU) | | Descriptor: | Cyclic (adenosine-(2'-5')-monophosphate-adenosine-(3'-5')-monophosphate, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FM1
 
 | |
8FMG
 
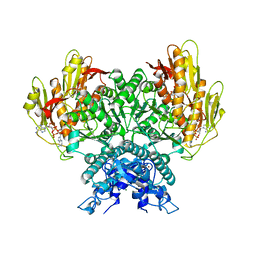 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-diAMP ligand (3 tetramers in the AU) | | Descriptor: | Cyclic (adenosine-(2'-5')-monophosphate-adenosine-(3'-5')-monophosphate, MAGNESIUM ION, SAVED domain-containing protein, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
