1IUU
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE AT PH 9.4 | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
3KIP
 
 | | Crystal structure of type-II 3-dehydroquinase from C. albicans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinase, type II, ... | | Authors: | Trapani, S, Schoehn, G, Navaza, J, Abergel, C. | | Deposit date: | 2009-11-02 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macromolecular crystal data phased by negative-stained electron-microscopy reconstructions.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1ZXE
 
 | | Crystal Structure of eIF2alpha Protein Kinase GCN2: D835N Inactivating Mutant in Apo Form | | Descriptor: | GLYCEROL, Serine/threonine-protein kinase | | Authors: | Padyana, A.K, Qiu, H, Roll-Mecak, A, Hinnebusch, A.G, Burley, S.K. | | Deposit date: | 2005-06-07 | | Release date: | 2005-06-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Autoinhibition and Mutational Activation of Eukaryotic Initiation Factor 2{alpha} Protein Kinase GCN2
J.Biol.Chem., 280, 2005
|
|
1J07
 
 | | Crystal structure of the mouse acetylcholinesterase-decidium complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,8-DIAMINO-5,10'-(TRIMETHYLAMMONIUM)DECYL-6-PHENYL PHENANTHRIDINIUM, CARBONATE ION, ... | | Authors: | Bourne, Y, Taylor, P, Radic, Z, Marchot, P. | | Deposit date: | 2002-11-07 | | Release date: | 2003-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site
EMBO J., 22, 2003
|
|
25C8
 
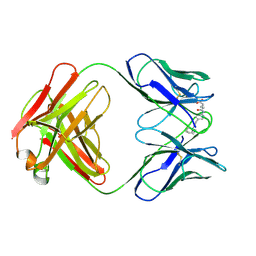 | | CATALYTIC ANTIBODY 5C8, FAB-HAPTEN COMPLEX | | Descriptor: | IGG 5C8, N-METHYL-N-(PARA-GLUTARAMIDOPHENYL-ETHYL)-PIPERIDINIUM ION | | Authors: | Gruber, K, Wilson, I.A. | | Deposit date: | 1998-03-18 | | Release date: | 1999-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antibody catalysis of a disfavored ring closure reaction.
Biochemistry, 38, 1999
|
|
1ZRL
 
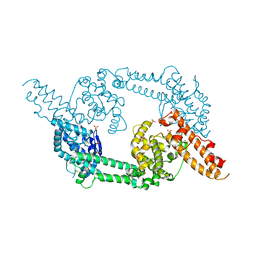 | | Crystal structure of EBA-175 Region II (RII) | | Descriptor: | CHLORIDE ION, SULFATE ION, erythrocyte binding antigen region II | | Authors: | Tolia, N.H, Enemark, E.J, Sim, B.K, Joshua-Tor, L. | | Deposit date: | 2005-05-19 | | Release date: | 2005-08-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the EBA-175 Erythrocyte Invasion Pathway of the Malaria Parasite Plasmodium falciparum.
Cell(Cambridge,Mass.), 122, 2005
|
|
1IKN
 
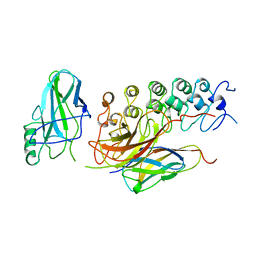 | | IKAPPABALPHA/NF-KAPPAB COMPLEX | | Descriptor: | PROTEIN (I-KAPPA-B-ALPHA), PROTEIN (NF-KAPPA-B P50D SUBUNIT), PROTEIN (NF-KAPPA-B P65 SUBUNIT) | | Authors: | Huxford, T, Huang, D.-B, Malek, S, Ghosh, G. | | Deposit date: | 1998-11-13 | | Release date: | 1999-04-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation.
Cell(Cambridge,Mass.), 95, 1998
|
|
1ZTK
 
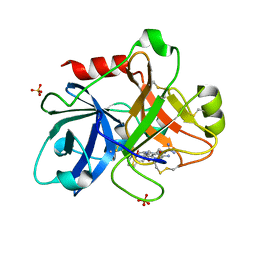 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 2-(5-Amino-6-oxo-2-m-tolyl-6H-pyrimidin-1-yl)-N-[4-guanidino-1-(thiazole-2-carbonyl)-butyl]-acetamide | | Descriptor: | 2-(5-AMINO-6-OXO-2-M-TOLYL-6H-PYRIMIDIN-1-YL)-N-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYL]-ACETAMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Nagafuji, P, Jin, L, Rynkiewicz, M, Quinn, J, Bibbins, F, Meyers, H, Babine, R.E, Strickler, J.E, Abdel-Meguid, S.S. | | Deposit date: | 2005-05-27 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyrimidinone Inhibitors of a Thrombolytic Protease
To be Published
|
|
1IN4
 
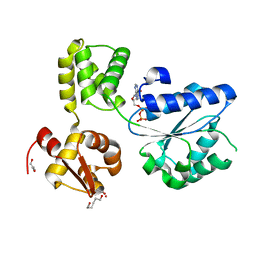 | |
1LBT
 
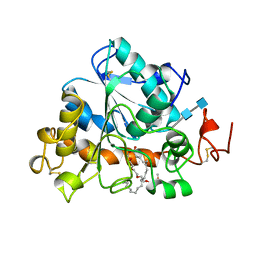 | | LIPASE (E.C.3.1.1.3) (TRIACYLGLYCEROL HYDROLASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LIPASE B, METHYLPENTA(OXYETHYL) HEPTADECANOATE | | Authors: | Uppenberg, J, Jones, T.A. | | Deposit date: | 1995-07-11 | | Release date: | 1995-10-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and molecular-modeling studies of lipase B from Candida antarctica reveal a stereospecificity pocket for secondary alcohols.
Biochemistry, 34, 1995
|
|
1LG2
 
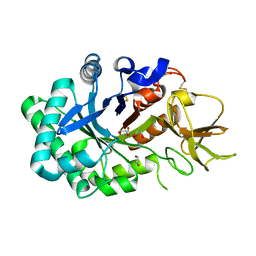 | |
2ACE
 
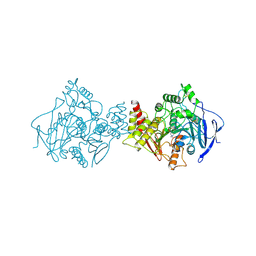 | | NATIVE ACETYLCHOLINESTERASE (E.C. 3.1.1.7) FROM TORPEDO CALIFORNICA | | Descriptor: | ACETYLCHOLINE, ACETYLCHOLINESTERASE | | Authors: | Harel, M, Raves, M.L, Silman, I, Sussman, J.L. | | Deposit date: | 1996-06-23 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of acetylcholinesterase complexed with the nootropic alkaloid, (-)-huperzine A.
Nat.Struct.Biol., 4, 1997
|
|
2AD7
 
 | | crystal structure of methanol dehydrogenase from M. W3A1 (form C) in the presence of methanol | | Descriptor: | CALCIUM ION, Methanol dehydrogenase subunit 1, Methanol dehydrogenase subunit 2, ... | | Authors: | Li, J, Gan, J.-H, Xia, Z.-X, Mathews, F.S. | | Deposit date: | 2005-07-20 | | Release date: | 2006-07-25 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Biochem.Biophys.Res.Commun., 406, 2011
|
|
2ADU
 
 | | Human Methionine Aminopeptidase Complex with 4-Aryl-1,2,3-triazole Inhibitor | | Descriptor: | 4-(3-METHYLPHENYL)-1H-1,2,3-TRIAZOLE, COBALT (II) ION, Methionine aminopeptidase 2 | | Authors: | Kallander, L.S, Lu, Q, Chen, W, Tomaszek, T, Yang, G, Tew, D, Meek, T.D, Hofmann, G.A, Schulz-Pritchard, C.K, Smith, W.W, Janson, C.A, Ryan, M.D, Zhang, G.F, Johanson, K.O, Kirkpatrick, R.B, Ho, T.F, Fisher, P.W, Mattern, M.R, Johnson, R.K, Hansbury, M.J, Winkler, J.D, Ward, K.W, Veber, D.F, Thompson, S.K. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 4-Aryl-1,2,3-triazole: A Novel Template for a Reversible Methionine Aminopeptidase 2 Inhibitor, Optimized To Inhibit Angiogenesis in Vivo
J.Med.Chem., 48, 2005
|
|
1LJF
 
 | |
1LJT
 
 | | Crystal Structure of Macrophage Migration Inhibitory Factor complexed with (S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole-acetic acid methyl ester (ISO-1) | | Descriptor: | 3-(4-HYDROXYPHENYL)-4,5-DIHYDRO-5-ISOXAZOLE-ACETIC ACID METHYL ESTER, Macrophage migration inhibitory factor | | Authors: | Lubetsky, J.B, Dios, A, Han, J, Aljabari, B, Ruzsicska, B, Mitchell, R, Lolis, E, Al-Abed, Y. | | Deposit date: | 2002-04-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Tautomerase Active Site of Macrophage Migration Inhibitory factor is a Potential Target for Discovery of Novel Anti-inflammatory Agents
J.Biol.Chem., 277, 2002
|
|
1LPA
 
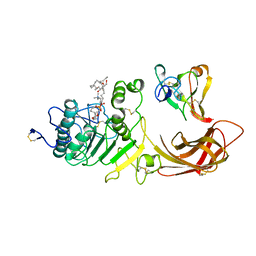 | | INTERFACIAL ACTIVATION OF THE LIPASE-PROCOLIPASE COMPLEX BY MIXED MICELLES REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, COLIPASE, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Van Tilbeurgh, H, Egloff, M.-P, Cambillau, C. | | Deposit date: | 1994-08-19 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by X-ray crystallography.
Nature, 362, 1993
|
|
1LKF
 
 | | LEUKOCIDIN F (HLGB) FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | LEUKOCIDIN F SUBUNIT | | Authors: | Olson, R, Nariya, H, Yokota, K, Kamio, Y, Gouaux, J.E. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel.
Nat.Struct.Biol., 6, 1999
|
|
1ZZC
 
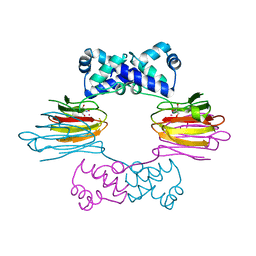 | | Crystal Structure of CoII HppE in Complex with Tris Buffer | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COBALT (II) ION, hydroxypropylphosphonic acid epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1LN1
 
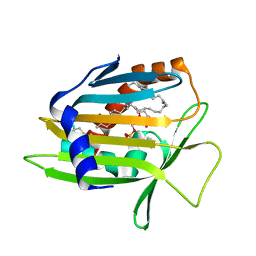 | | Crystal Structure of Human Phosphatidylcholine Transfer Protein in Complex with Dilinoleoylphosphatidylcholine | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylcholine transfer protein | | Authors: | Roderick, S.L, Chan, W.W, Agate, D.S, Olsen, L.R, Vetting, M.W, Rajashankar, K.R, Cohen, D.E. | | Deposit date: | 2002-05-02 | | Release date: | 2002-06-26 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human phosphatidylcholine transfer protein in complex with its ligand.
Nat.Struct.Biol., 9, 2002
|
|
1ZYL
 
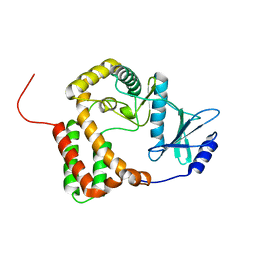 | |
1LOF
 
 | |
208D
 
 | | HIGH-RESOLUTION STRUCTURE OF A DNA HELIX FORMING (C.G)*G BASE TRIPLETS | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*AP*TP*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Van Meervelt, L, Vlieghe, D, Dautant, A, Gallois, B, Precigoux, G, Kennard, O. | | Deposit date: | 1995-04-26 | | Release date: | 1995-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-resolution structure of a DNA helix forming (C.G)*G base triplets.
Nature, 374, 1995
|
|
243D
 
 | | STRUCTURE OF THE DNA OCTANUCLEOTIDE D(ACGTACGT)2 | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*AP*CP*GP*T)-3') | | Authors: | Wilcock, D.J, Adams, A, Cardin, C.J, Wakelin, L.P.G. | | Deposit date: | 1996-01-10 | | Release date: | 1996-02-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DNA octanucleotide d(ACGTACGT)2.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1LUE
 
 | | RECOMBINANT SPERM WHALE MYOGLOBIN H64D/V68A/D122N MUTANT (MET) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2002-05-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular engineering of myoglobin: influence of residue 68 on the rate and the
enantioselectivity of oxidation reactions catalyzed by H64D/V68X myoglobin
Biochemistry, 42, 2003
|
|
