8AJB
 
 | |
7XHT
 
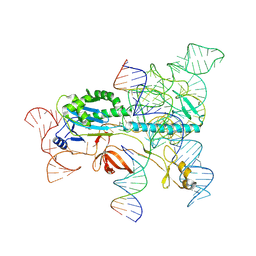 | | Structure of the OgeuIscB-omega RNA-target DNA complex | | Descriptor: | DNA (49-MER), DNA (5'-D(P*GP*AP*AP*GP*AP*AP*AP*AP*CP*CP*AP*T)-3'), LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Kato, K, Okazaki, O, Isayama, Y, Ishikawa, J, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-04-10 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structure of the IscB-omega RNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9.
Nat Commun, 13, 2022
|
|
7WY4
 
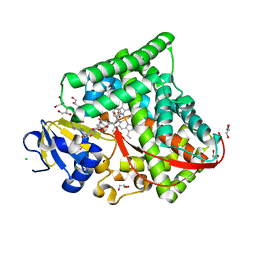 | | Structure of the CYP102A1 F87A Haem Domain with N-Enanthyl-L-Prolyl-L-Phenylalanine in complex with Styrene | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | Authors: | Suzuki, K, Stanfield, J.K, Shisaka, Y, Omura, K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2022-02-15 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Compound I Mimic Reveals the Transient Active Species of a Cytochrome P450 Enzyme: Insight into the Stereoselectivity of P450-Catalysed Oxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WY3
 
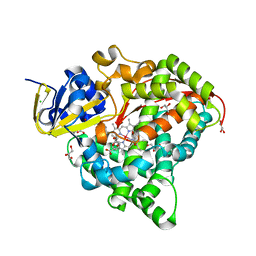 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 F87V Mutant Haem Domain with N-(5-Cyclohexyl)valeroyl-L-Phenylalanine in complex with Styrene | | Descriptor: | (2~{S})-2-(5-cyclohexylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | Authors: | Suzuki, K, Stanfield, J.K, Shisaka, Y, Omura, K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2022-02-15 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Compound I Mimic Reveals the Transient Active Species of a Cytochrome P450 Enzyme: Insight into the Stereoselectivity of P450-Catalysed Oxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WY2
 
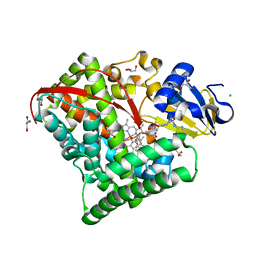 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 F87A Mutant Haem Domain with N-Enanthyl-L-Prolyl-L-Phenylalanine in complex with Styrene | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | Authors: | Suzuki, K, Stanfield, J.K, Shisaka, Y, Omura, K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2022-02-15 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Compound I Mimic Reveals the Transient Active Species of a Cytochrome P450 Enzyme: Insight into the Stereoselectivity of P450-Catalysed Oxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WY1
 
 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Enanthyl-L-Prolyl-L-Phenylalanine in complex with Styerene | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, Oxomolybdenum Mesoporphyrin IX, ... | | Authors: | Suzuki, K, Stanfield, J.K, Shisaka, Y, Omura, K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2022-02-15 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Compound I Mimic Reveals the Transient Active Species of a Cytochrome P450 Enzyme: Insight into the Stereoselectivity of P450-Catalysed Oxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8AHW
 
 | | Structure of DCS-resistant variant D322N of alanine racemase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Alanine racemase, GLYCEROL | | Authors: | de Chiara, C, Prosser, G, Ogrodowicz, R.W, de Carvalho, L.P.S. | | Deposit date: | 2022-07-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the d-Cycloserine-Resistant Variant D322N of Alanine Racemase from Mycobacterium tuberculosis .
Acs Bio Med Chem Au, 3, 2023
|
|
8AVM
 
 | | Mutant of Superoxide Dismutase sodfm2 from Bacteroides fragilis | | Descriptor: | FE (III) ION, Superoxide dismutase [Fe] | | Authors: | Basle, A, Barwinska-Sendra, A, Sendra, K.M, Waldron, K. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An ancient metalloenzyme evolves through metal preference modulation.
Nat Ecol Evol, 7, 2023
|
|
7W62
 
 | | Mevo lectin complex with mannotetraose (Man4) | | Descriptor: | 1,2-ETHANEDIOL, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Sivaji, N, Vijayan, M. | | Deposit date: | 2021-12-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mevo lectin complex with mannotetraose (Man4)
To Be Published
|
|
7ZBH
 
 | |
8AMS
 
 | | Complex of human TRIM2 RING domain, UBCH5C, and Ubiquitin | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Polyubiquitin-C, ... | | Authors: | Perez-Borrajero, C, Kotova, I, Murciano, B, Hennig, J. | | Deposit date: | 2022-08-04 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
8BOT
 
 | |
7WZ3
 
 | | Cryo-EM structure of human TRiC-tubulin-S1 state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM.
Commun Biol, 6, 2023
|
|
8ABY
 
 | |
8AC0
 
 | |
8AD1
 
 | |
8ACP
 
 | |
8ABZ
 
 | |
8AC2
 
 | | RNA polymerase- post-terminated, open clamp state | | Descriptor: | DNA Non-template strand, DNA Template strand, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Dey, S, Weixlbaumer, A. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into RNA-mediated transcription regulation in bacteria.
Mol.Cell, 82, 2022
|
|
8A9J
 
 | |
8AC1
 
 | |
7XL4
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (closed lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
8A9K
 
 | | Cryo-EM structure of USP1-UAF1 bound to FANCI and mono-ubiquitinated FANCD2 with ML323 (consensus reconstruction) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, DNA (61-MER), Fanconi anemia group D2 protein, ... | | Authors: | Rennie, M.L, Walden, H. | | Deposit date: | 2022-06-28 | | Release date: | 2022-10-12 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site.
Sci Adv, 8, 2022
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | Descriptor: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XL3
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (open lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
