8RGR
 
 | | Closed Complex I from murine liver | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-12-14 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QQS
 
 | |
8D39
 
 | |
5TS3
 
 | |
8QQK
 
 | | Cryo-EM structure of E. coli cytochrome bo3 quinol oxidase assembled in peptidiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, ... | | Authors: | Gao, Y, Zhang, Y, Hakke, S, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2023-10-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of cytochrome bo 3 quinol oxidase assembled in peptidiscs reveals an "open" conformation for potential ubiquinone-8 release.
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
8JYG
 
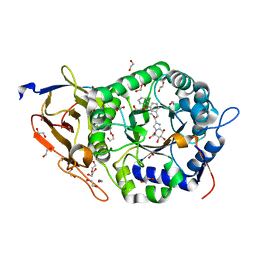 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-2-[2-(3-phenoxyphenyl)ethyl]-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based lead optimization to improve potency and selectivity of a novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid series of heparanase-1 inhibitor.
Bioorg.Med.Chem., 93, 2023
|
|
1AHD
 
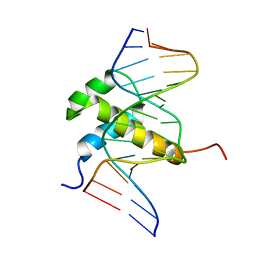 | | DETERMINATION OF THE NMR SOLUTION STRUCTURE OF AN ANTENNAPEDIA HOMEODOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), Homeotic protein antennapedia | | Authors: | Billeter, M, Qian, Y.Q, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex.
J.Mol.Biol., 234, 1993
|
|
5TTY
 
 | | PagF prenyltransferase | | Descriptor: | DI(HYDROXYETHYL)ETHER, PagF prenyltransferase | | Authors: | Hao, Y, Nair, S.K. | | Deposit date: | 2016-11-04 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the broad substrate selectivity of a peptide prenyltransferase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7ZDM
 
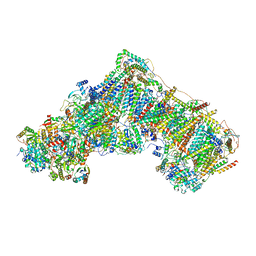 | | Complex I from Ovis aries at pH5.5, Closed state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7JY9
 
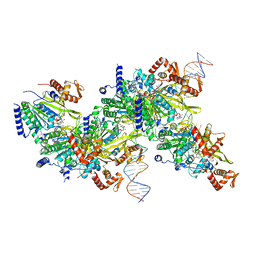 | | Structure of a 9 base pair RecA-D loop complex | | Descriptor: | DNA (27-MER), DNA (42-MER), MAGNESIUM ION, ... | | Authors: | Pavletich, N.P. | | Deposit date: | 2020-08-29 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of strand exchange from RecA-DNA synaptic and D-loop structures.
Nature, 586, 2020
|
|
8D59
 
 | | Crystal structure of human METTL1 in complex with SAM | | Descriptor: | CACODYLATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raj, R, Babu, K, Nam, Y. | | Deposit date: | 2022-06-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures and mechanisms of tRNA methylation by METTL1-WDR4.
Nature, 613, 2023
|
|
8D5B
 
 | | Crystal structure of human METTL1 in complex with SAH | | Descriptor: | CACODYLATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raj, R, Babu, K, Nam, Y. | | Deposit date: | 2022-06-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures and mechanisms of tRNA methylation by METTL1-WDR4.
Nature, 613, 2023
|
|
5A3U
 
 | | HIF prolyl hydroxylase 2 (PHD2/EGLN1) in complex with 6-(5-oxo-4-(1H- 1,2,3-triazol-1-yl)-2,5-dihydro-1H-pyrazol-1-yl)nicotinic acid | | Descriptor: | 6-(5-oxo-4-(1H-1,2,3-triazol-1-yl)-2,5-dihydro-1H-pyrazol-1-yl)nicotinic acid, EGL NINE HOMOLOG 1, MANGANESE (II) ION | | Authors: | Chowdhury, R, Gomez-Perez, V, Schofield, C.J. | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Potent and Selective Triazole-Based Inhibitors of the Hypoxia-Inducible Factor Prolyl-Hydroxylases with Activity in the Murine Brain.
Plos One, 10, 2015
|
|
7K4Z
 
 | | Crystal structure of Kemp Eliminase HG3.17 in complex with the transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, PENTAETHYLENE GLYCOL | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7ZEB
 
 | | Complex I from Ovis aries at pH9, Closed state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-30 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
8QNR
 
 | | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: G413N variant in complex with L-gulono-1,4-lactone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-galactono-1,4-lactone dehydrogenase, L-gulono-1,4-lactone | | Authors: | Boverio, A, Mattevi, A. | | Deposit date: | 2023-09-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: G413N variant in complex with L-gulono-1,4-lactone
To Be Published
|
|
7ZDP
 
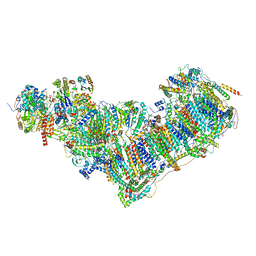 | | Complex I from Ovis aries at pH9, Open state | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE MONOPHOSPHATE, Acyl carrier protein, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
8D9L
 
 | |
5AEC
 
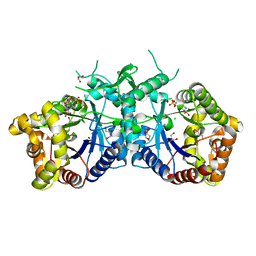 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7K4Q
 
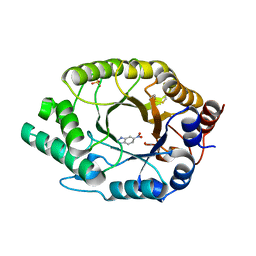 | | Crystal structure of Kemp Eliminase HG3 in complex with the transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
6VCA
 
 | | TB38 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, ... | | Authors: | Zhou, Y.F, Lord, D.M. | | Deposit date: | 2019-12-20 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | A highly potent CD73 biparatopic antibody blocks organization of the enzyme active site through dual mechanisms.
J.Biol.Chem., 295, 2020
|
|
7K4V
 
 | | Crystal structure of Kemp Eliminase HG3.17 | | Descriptor: | Endo-1,4-beta-xylanase, TETRAETHYLENE GLYCOL | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
8QMY
 
 | |
1I4J
 
 | | CRYSTAL STRUCTURE OF L22 RIBOSOMAL PROTEIN MUTANT | | Descriptor: | 50S RIBOSOMAL PROTEIN L22 | | Authors: | Davydova, N.L, Streltsov, V.A, Fedorov, R, Wilce, M, Liljas, A, Garder, M. | | Deposit date: | 2001-02-22 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | L22 ribosomal protein and effect of its mutation on ribosome resistance to erythromycin.
J.Mol.Biol., 322, 2002
|
|
6VDE
 
 | | Full-length M. smegmatis Pol1 | | Descriptor: | DNA polymerase I, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
