6K18
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mn | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6K1A
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mn and Mg | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MAGNESIUM ION, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
8K38
 
 | | The structure of bacteriophage lambda portal-adaptor | | Descriptor: | Head completion protein, Portal protein B | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K39
 
 | | Structure of the bacteriophage lambda portal vertex | | Descriptor: | Major capsid protein, Portal protein B | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
7BI0
 
 | | GA repetition with i-motif clip at 5'-end | | Descriptor: | (CH+)C(CH+)GAGA, C(CH+)CGAGA | | Authors: | Novotny, A, Novotny, J. | | Deposit date: | 2021-01-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing structural peculiarities of homopurine GA repetition stuck by i-motif clip.
Nucleic Acids Res., 49, 2021
|
|
7BL0
 
 | | GA attached to an i-motif clip at 3'-end | | Descriptor: | GA(CH+)C(5MeCH+), GAC(HCY)(5MeC) | | Authors: | Novotny, A, Novotny, J. | | Deposit date: | 2021-01-17 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing structural peculiarities of homopurine GA repetition stuck by i-motif clip.
Nucleic Acids Res., 49, 2021
|
|
7BLM
 
 | |
7BMA
 
 | |
3BIP
 
 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | FACT complex subunit SPT16 | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
1NVQ
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/UCN-01 | | Descriptor: | 7-HYDROXYSTAUROSPORINE, Peptide ASVSA, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
7B2X
 
 | | Crystal structure of human 5' exonuclease Appollo H61Y variant | | Descriptor: | 5' exonuclease Apollo, NICKEL (II) ION | | Authors: | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
1NVR
 
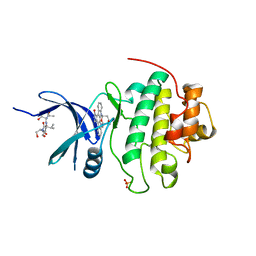 | | The Complex Structure Of Checkpoint Kinase Chk1/Staurosporine | | Descriptor: | Peptide ASVSA, STAUROSPORINE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
2O9A
 
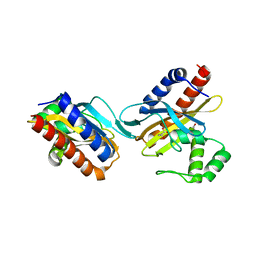 | | The crystal structure of the E.coli IclR C-terminal fragment in complex with pyruvate. | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, PYRUVIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
7AKR
 
 | |
7AKS
 
 | |
7ARW
 
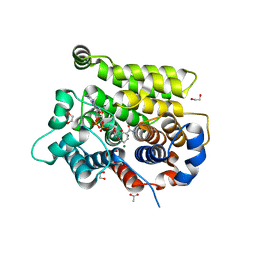 | | Structure of human ARH3 E41A bound to alpha-NAD+ and magnesium | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADP-ribose glycohydrolase ARH3, ... | | Authors: | Rack, J.G.M, Zorzini, V, Ahel, I. | | Deposit date: | 2020-10-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Nat Commun, 12, 2021
|
|
6JVK
 
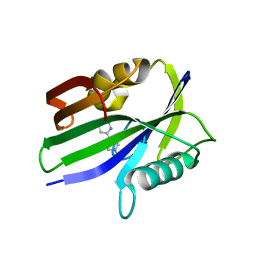 | | Crystal structure of human MTH1 in complex with compound MI1012 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-(4-methylpiperazin-1-yl)pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVT
 
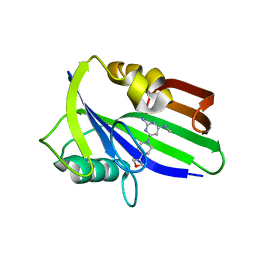 | | Crystal structure of human MTH1 in complex with compound MI1030 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-[4-(oxetan-3-yl)piperazin-1-yl]pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
1XO0
 
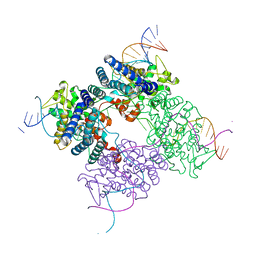 | | High resolution structure of the holliday junction intermediate in cre-loxp site-specific recombination | | Descriptor: | Recombinase CRE, loxP | | Authors: | Ghosh, K, Lau, C.K, Guo, F, Segall, A.M, Van Duyne, G.D. | | Deposit date: | 2004-10-05 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination.
J.Biol.Chem., 280, 2005
|
|
2O99
 
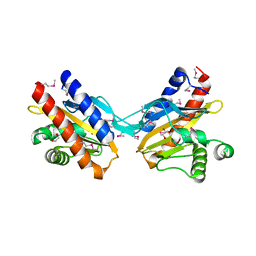 | | The crystal structure of E.coli IclR C-terminal fragment in complex with glyoxylate | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, GLYCOLIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
3SXK
 
 | |
3DBV
 
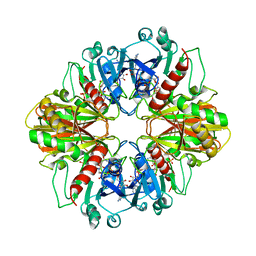 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1997-01-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
6JVJ
 
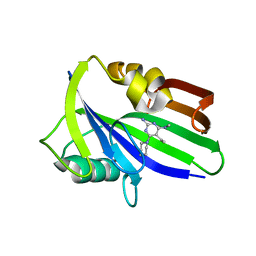 | | Crystal structure of human MTH1 in complex with compound MI1006 | | Descriptor: | 5-ethyl-N4-methyl-6-piperidin-1-yl-pyrimidine-2,4-diamine, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVR
 
 | | Crystal structure of human MTH1 in complex with compound MI1026 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6K19
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mg | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MAGNESIUM ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
