2KN0
 
 | | Solution NMR Structure of xenopus Fn14 | | Descriptor: | Fn14 | | Authors: | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | Deposit date: | 2009-08-11 | | Release date: | 2011-06-29 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
3E9L
 
 | | Crystal Structure of Human Prp8, Residues 1755-2016 | | Descriptor: | CHLORIDE ION, Pre-mRNA-processing-splicing factor 8, SODIUM ION | | Authors: | Pena, V, Rozov, A, Wahl, M.C. | | Deposit date: | 2008-08-22 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of an RNase H domain at the heart of the spliceosome.
Embo J., 27, 2008
|
|
3DB4
 
 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, SULFATE ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-09-16 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
3E9O
 
 | |
3E9P
 
 | |
3DKR
 
 | |
3DYI
 
 | |
3E1R
 
 | | Midbody targeting of the ESCRT machinery by a non-canonical coiled-coil in CEP55 | | Descriptor: | Centrosomal protein of 55 kDa, Programmed cell death 6-interacting protein | | Authors: | Lee, H.H, Elia, N, Ghirlando, R, Lippincott-Schwartz, J, Hurley, J.H. | | Deposit date: | 2008-08-04 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55.
Science, 322, 2008
|
|
3E66
 
 | | Crystal structure of the beta-finger domain of yeast Prp8 | | Descriptor: | PRP8 | | Authors: | Yang, K, Zhang, L, Xu, T, Heroux, A, Zhao, R. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3FDQ
 
 | |
3DB3
 
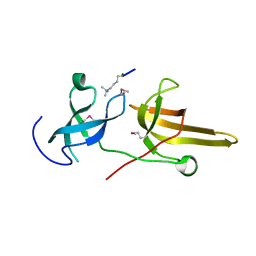 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 in complex with trimethylated histone H3-K9 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Trimethylated histone H3-K9 peptide | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-09-16 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
3DYV
 
 | |
3F2E
 
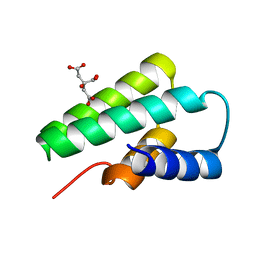 | | Crystal structure of Yellowstone SIRV coat protein C-terminus | | Descriptor: | CITRIC ACID, SIRV coat protein | | Authors: | Taurog, R.E, Szymczyna, B.R, Williamson, J.R, Johnson, J.E. | | Deposit date: | 2008-10-29 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | Synergy of NMR, computation, and X-ray crystallography for structural biology.
Structure, 17, 2009
|
|
3E1G
 
 | |
3DLT
 
 | |
3FZE
 
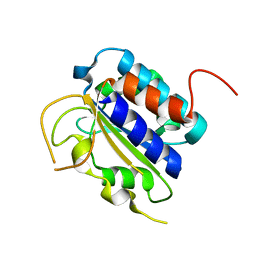 | | Structure of the 'minimal scaffold' (ms) domain of Ste5 that cocatalyzes Fus3 phosphorylation by Ste7 | | Descriptor: | Protein STE5 | | Authors: | Good, M.C, Tang, G, Singleton, J, Remenyi, A, Lim, W.A. | | Deposit date: | 2009-01-25 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation.
Cell(Cambridge,Mass.), 136, 2009
|
|
3ENB
 
 | |
3FRL
 
 | | The 2.25 A crystal structure of LipL32, the major surface antigen of Leptospira interrogans serovar Copenhageni | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, CHLORIDE ION, LipL32, ... | | Authors: | Farah, C.S, Guzzo, C.R, Hauk, P, Ho, P.L. | | Deposit date: | 2009-01-08 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and calcium-binding activity of LipL32, the major surface antigen of pathogenic Leptospira sp.
J.Mol.Biol., 390, 2009
|
|
3FNB
 
 | | Crystal structure of acylaminoacyl peptidase SMU_737 from Streptococcus mutans UA159 | | Descriptor: | 1,2-ETHANEDIOL, Acylaminoacyl peptidase SMU_737, BETA-MERCAPTOETHANOL, ... | | Authors: | Kim, Y, Hatzos, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-23 | | Release date: | 2009-01-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1178 Å) | | Cite: | Crystal structure of acylaminoacyl-peptidase SMU_737 from Streptococcus mutans UA159
To be Published
|
|
3G74
 
 | | Crystal structure of a functionally unknown protein from Eubacterium ventriosum ATCC 27560 | | Descriptor: | Protein of unknown function, SULFATE ION | | Authors: | Tan, K, Sather, A, Marshall, N, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of a functionally unknown protein from Eubacterium ventriosum ATCC 27560
To be Published
|
|
5XNG
 
 | | EFK17A structure in Microgel MAA60 | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2017-05-22 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational Aspects of High Content Packing of Antimicrobial Peptides in Polymer Microgels
ACS Appl Mater Interfaces, 9, 2017
|
|
5XVL
 
 | | Crystal structure of AL2 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 2, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
5XRX
 
 | | EFK17DA structure in Microgel MAA60 | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2017-06-10 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational Aspects of High Content Packing of Antimicrobial Peptides in Polymer Microgels
ACS Appl Mater Interfaces, 9, 2017
|
|
5Y53
 
 | |
5ZUN
 
 | | Crystal structure of human monoacylglycerol lipase in complex with compound 3l | | Descriptor: | (4R)-1-(2'-chloro[1,1'-biphenyl]-3-yl)-4-[4-(1,3-thiazole-2-carbonyl)piperazin-1-yl]pyrrolidin-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Sogabe, S, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, Synthesis, and Evaluation of Piperazinyl Pyrrolidin-2-ones as a Novel Series of Reversible Monoacylglycerol Lipase Inhibitors
J. Med. Chem., 61, 2018
|
|
