5LHI
 
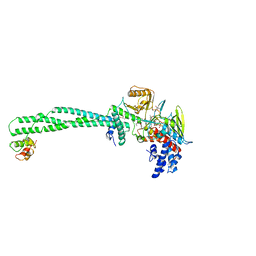 | | Structure of the KDM1A/CoREST complex with the inhibitor N-[3-(ethoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]-4-methylthieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-12 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
5LHH
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor 4-ethyl-N-[3-(methoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | 4-ethyl-~{N}-[3-(methoxymethyl)-2-[[4-[[(3~{R})-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
5LHG
 
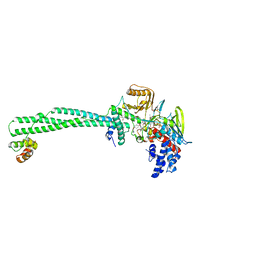 | | Structure of the KDM1A/CoREST complex with the inhibitor 4-methyl-N-[4-[[4-(1-methylpiperidin-4-yl)oxyphenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | 4-methyl-N-[4-[[4-[(1-methyl-4-piperidyl)oxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
2GFA
 
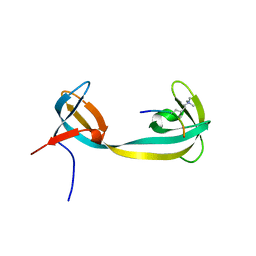 | | double tudor domain complex structure | | Descriptor: | Jumonji domain-containing protein 2A, peptide | | Authors: | Huang, Y, Fang, J, Bedford, M.T, Zhang, Y, Xu, R.M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A
Science, 312, 2006
|
|
2GF7
 
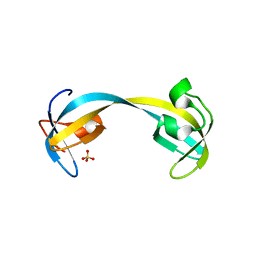 | | Double tudor domain structure | | Descriptor: | Jumonji domain-containing protein 2A, SULFATE ION | | Authors: | Huang, Y, Fang, J, Bedford, M.T, Zhang, Y, Xu, R.M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A
Science, 312, 2006
|
|
3QLN
 
 | | Crystal structure of ATRX ADD domain in free state | | Descriptor: | Transcriptional regulator ATRX, ZINC ION | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QLA
 
 | | Hexagonal complex structure of ATRX ADD bound to H3K9me3 peptide | | Descriptor: | POTASSIUM ION, Transcriptional regulator ATRX, ZINC ION, ... | | Authors: | Xiang, B, Li, H. | | Deposit date: | 2011-02-02 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QZT
 
 | | Crystal Structure of BPTF bromo in complex with histone H4K16ac - Form II | | Descriptor: | GLYCEROL, Histone H4, Nucleosome-remodeling factor subunit BPTF | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QZS
 
 | |
3QZV
 
 | | Crystal Structure of BPTF PHD-linker-bromo in complex with histone H4K12ac peptide | | Descriptor: | Histone H4, Nucleosome-remodeling factor subunit BPTF, ZINC ION | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
4LKA
 
 | | Crystal Structure of MOZ double PHD finger histone H3K9ac complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-07 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
3SMR
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Wasney, G.A, Tempel, W, Senisterra, G, Bolshan, Y, Smil, D, Nguyen, K.T, Hajian, T, Poda, G, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
4LLB
 
 | | Crystal Structure of MOZ double PHD finger histone H3K14ac complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
4LJN
 
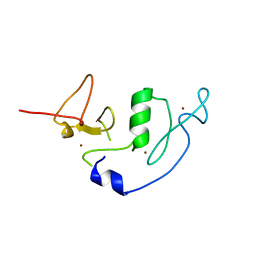 | | Crystal Structure of MOZ double PHD finger | | Descriptor: | Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
4LK9
 
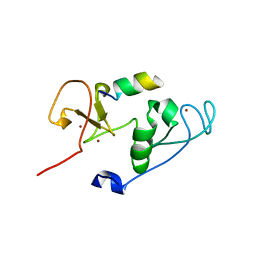 | | Crystal Structure of MOZ double PHD finger histone H3 tail complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-07 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
3QL9
 
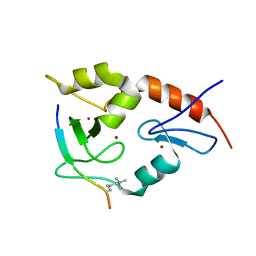 | |
3QLC
 
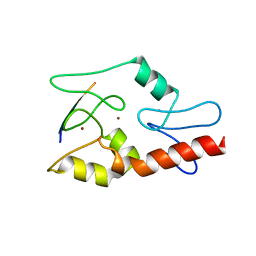 | |
5A1H
 
 | | Crystal structure of human Spindlin3 | | Descriptor: | SPINDLIN-3 | | Authors: | Srikannathasan, V, Gileadi, C, Johansson, C, Shrestha, L, Tallon, R, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2015-04-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Spindlin3
To be Published
|
|
5A3P
 
 | | Crystal structure of the catalytic domain of human PLU1 (JARID1B). | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Tallant, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5A3W
 
 | | Crystal structure of human PLU-1 (JARID1B) in complex with Pyridine-2, 6-dicarboxylic Acid (PDCA) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Kopec, J, Strain-Damerell, C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5CEH
 
 | | Structure of histone lysine demethylase KDM5A in complex with selective inhibitor | | Descriptor: | 7-oxo-5-phenyl-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, NICKEL (II) ION, ... | | Authors: | Kiefer, J.R, Vinogradova, M. | | Deposit date: | 2015-07-06 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells.
Nat.Chem.Biol., 12, 2016
|
|
7D4A
 
 | | The Crystal Structure of human JMJD2A Tudor domain from Biortus | | Descriptor: | Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Ju, C, Bao, X, Zhu, B. | | Deposit date: | 2020-09-23 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | The Crystal Structure of human JMJD2A from Biortus.
To Be Published
|
|
5A1F
 
 | | Crystal structure of the catalytic domain of PLU1 in complex with N-oxalylglycine. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Srikannathasan, V, Johansson, C, Strain-Damerell, C, Gileadi, C, Szykowska, A, Kupinska, K, Kopec, J, Krojer, T, Steuber, H, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5D6W
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A | | Descriptor: | Lysine-specific demethylase 4A, S,R MESO-TARTARIC ACID | | Authors: | Wang, F, Su, Z, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
5A3T
 
 | | Crystal structure of human PLU-1 (JARID1B) in complex with KDM5-C49 (2-(((2-((2-(dimethylamino)ethyl)(ethyl)amino)-2-oxoethyl)amino)methyl) isonicotinic acid). | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Kopec, J, Strain-Damerell, C, Kupinska, K, BurgessBrown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-06-17 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
