1Z3M
 
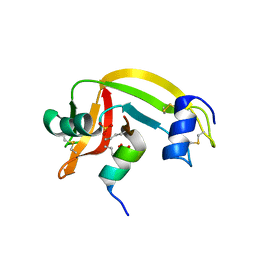 | | Crystal structure of mutant Ribonuclease S (F8Nva) | | Descriptor: | Ribonuclease pancreatic, S-Peptide, S-protein, ... | | Authors: | Das, M, Vasudeva Rao, B, Ghosh, S, Varadarajan, R. | | Deposit date: | 2005-03-14 | | Release date: | 2005-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Attempts to delineate the relative contributions of changes in hydrophobicity and packing to changes in stability of ribonuclease S mutants.
Biochemistry, 44, 2005
|
|
4DV5
 
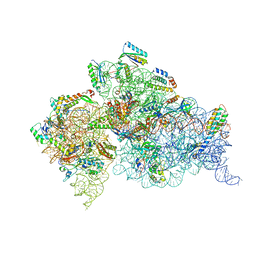 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A914G, bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.683 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
2BQR
 
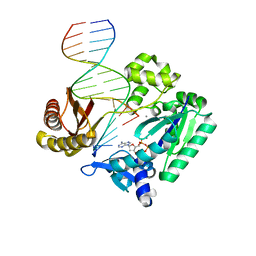 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*TP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-27 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
8QRL
 
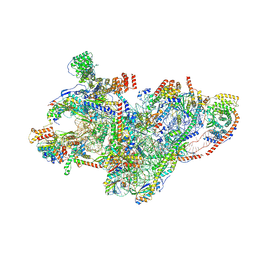 | | mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 2 | | Descriptor: | 12S mitochondrial rRNA, 12S rRNA N4-methylcytidine (m4C) methyltransferase, 28S ribosomal protein S10, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-09 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
4DUZ
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U13C, bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.651 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DUY
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U13C | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.386 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DV4
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A914G | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.651 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
1Z3P
 
 | | X-Ray crystal structure of a mutant Ribonuclease S (M13Nva) | | Descriptor: | Ribonuclease pancreatic, S-Peptide, S-Protein, ... | | Authors: | Das, M, Rao, B.V, Ghosh, S, Varadarajan, R. | | Deposit date: | 2005-03-14 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Attempts to delineate the relative contributions of changes in hydrophobicity and packing to changes in stability of ribonuclease S mutants.
Biochemistry, 44, 2005
|
|
6BHJ
 
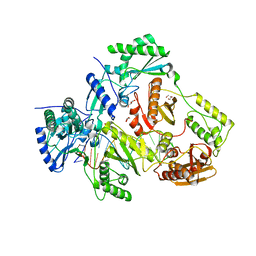 | | Structure of HIV-1 Reverse Transcriptase Bound to a 38-mer Hairpin Template-Primer RNA-DNA Aptamer | | Descriptor: | 38-MER RNA-DNA Aptamer, GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Ruiz, F.X, Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Integrative Structural Biology Studies of HIV-1 Reverse Transcriptase Binding to a High-Affinity DNA Aptamer
Curr Res Struct Biol, 2020
|
|
6W6P
 
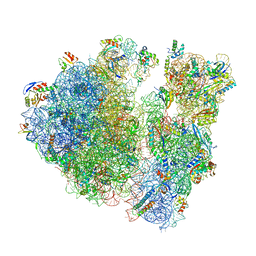 | |
4DV2
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, C912A | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.646 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
3Q0Z
 
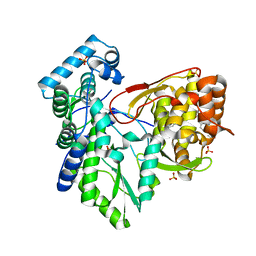 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5h-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Sheriff, S. | | Deposit date: | 2010-12-16 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Syntheses and initial evaluation of a series of indolo-fused heterocyclic inhibitors of the polymerase enzyme (NS5B) of the hepatitis C virus.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7LHX
 
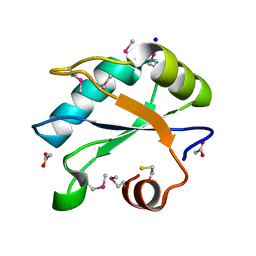 | | Human U1A protein with F37M and F77M mutations for improved phasing | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, SODIUM ION, ... | | Authors: | Jenkins, J.L, Lippa, G.M, Wedekind, J.E. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Affinity and Structural Analysis of the U1A RNA Recognition Motif with Engineered Methionines to Improve Experimental Phasing
Crystals, 11, 2021
|
|
4DV3
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, C912A, bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.547 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
7Z4D
 
 | |
3QGF
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(6-chloropyridazin-3-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(6-chloropyridazin-3-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QGG
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and N-cyclopropyl-6-[(3R)-3-{[4-(trifluoromethoxy)benzyl]carbamoyl}-4-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazin-1-yl]pyridazine-3-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, N-cyclopropyl-6-[(3R)-3-{[4-(trifluoromethoxy)benzyl]carbamoyl}-4-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazin-1-yl]pyridazine-3-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2QEX
 
 | |
5L3T
 
 | | Structure of the Saccharomyces cerevisiae TREX-2 complex | | Descriptor: | 26S proteasome complex subunit SEM1, Nuclear mRNA export protein SAC3, Nuclear mRNA export protein THP1, ... | | Authors: | Stewart, M, Aibara, S. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.927 Å) | | Cite: | The Sac3 TPR-like region in the Saccharomyces cerevisiae TREX-2 complex is more extensive but independent of the CID region.
J. Struct. Biol., 195, 2016
|
|
3LVP
 
 | | Crystal structure of bisphosphorylated IGF1-R Kinase domain (2P) in complex with a bis-azaindole inhibitor | | Descriptor: | 3-(4-chloro-1H-pyrrolo[2,3-b]pyridin-2-yl)-5,6-dimethoxy-1-methyl-1H-pyrrolo[3,2-b]pyridine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Insulin-like growth factor 1 receptor, ... | | Authors: | Maignan, S, Marquette, J.P, Guilloteau, J.P. | | Deposit date: | 2010-02-22 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design of Potent IGF1-R Inhibitors Related to Bis-azaindoles
Chem.Biol.Drug Des., 76, 2010
|
|
3VKW
 
 | | Crystal Structure of the Superfamily 1 Helicase from Tomato Mosaic Virus | | Descriptor: | Replicase large subunit, SULFATE ION | | Authors: | Nishikiori, M, Sugiyama, S, Xiang, H, Niiyama, M, Ishibashi, K, Inoue, T, Ishikawa, M, Matsumura, H, Katoh, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superfamily 1 helicase from tomato mosaic virus
J.Virol., 86, 2012
|
|
8FLK
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | | Descriptor: | 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
7KE0
 
 | |
4L69
 
 | |
