7Q81
 
 | |
6I01
 
 | | Structure of human D-glucuronyl C5 epimerase in complex with substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6CKR
 
 | | Crystal Structure of BRD4 with QC4956 | | Descriptor: | Bromodomain-containing protein 4, N-{3-[2-methyl-6-(1-methyl-1H-pyrazol-4-yl)-1-oxo-1,2-dihydroisoquinolin-4-yl]phenyl}methanesulfonamide | | Authors: | Hosfield, D.J. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis and biological evaluation of novel 4-phenylisoquinolinone BET bromodomain inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6I0C
 
 | | Human butyrylcholinesterase in complex with the R enantiomer of a chlorotacrine-tryptophan multi-target inhibitor. | | Descriptor: | (2~{R})-2-azanyl-~{N}-[6-[(6-chloranyl-1,2,3,4-tetrahydroacridin-9-yl)amino]hexyl]-3-(1~{H}-indol-3-yl)propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Nachon, F. | | Deposit date: | 2018-10-25 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.675 Å) | | Cite: | Novel tacrine-tryptophan hybrids: Multi-target directed ligands as potential treatment for Alzheimer's disease.
Eur.J.Med.Chem., 168, 2019
|
|
6YQ0
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3-oxidanyl-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6FG0
 
 | | Crystal structure of R. ruber ADH-A, mutant Y54G, F43T, L119Y, F282W | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2018-01-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Stereo- and Regioselectivity in Catalyzed Transformation of a 1,2-Disubstituted Vicinal Diol and the Corresponding Diketone by Wild Type and Laboratory Evolved Alcohol Dehydrogenases
Acs Catalysis, 8, 2018
|
|
8AID
 
 | |
6YNH
 
 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 4 of 4 | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
6I0N
 
 | |
6YRB
 
 | | Crystal structure of the tetramerization domain of the glycoprotein Gn (Andes virus) at pH 7.5 | | Descriptor: | Envelope polyprotein, IODIDE ION, RNA (5'-D(*())-R(P*UP*UP*UP*())-3'), ... | | Authors: | Serris, A, Rey, F.A, Guardado-Calvo, P. | | Deposit date: | 2020-04-20 | | Release date: | 2020-10-14 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | The Hantavirus Surface Glycoprotein Lattice and Its Fusion Control Mechanism.
Cell, 183, 2020
|
|
6Q9P
 
 | | Crystal structure of human Arginase-1 at pH 9.0 in complex with ABH | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase-1, MANGANESE (II) ION, ... | | Authors: | Grobben, Y, Uitdehaag, J.C.M, Zaman, G.J.R. | | Deposit date: | 2018-12-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural insights into human Arginase-1 pH dependence and its inhibition by the small molecule inhibitor CB-1158.
J Struct Biol X, 4, 2020
|
|
8B10
 
 | | Crystal Structure of Shank2-SAM mutant domain - L1800W | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bento, I, Gracia Alai, M, Kreienkamp, J.-H. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural deficits in key domains of Shank2 lead to alterations in postsynaptic nanoclusters and to a neurodevelopmental disorder in humans.
Mol Psychiatry, 2022
|
|
6H88
 
 | | MamM CTD D249N | | Descriptor: | BETA-MERCAPTOETHANOL, Magnetosome protein MamM, Cation efflux protein family | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2018-08-02 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | MamM CTD D249N
To Be Published
|
|
8ATJ
 
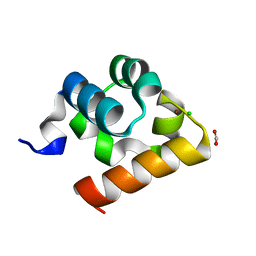 | | Crystal Structure of Shank2-SAM domain | | Descriptor: | CHLORIDE ION, FORMIC ACID, Isoform 4 of SH3 and multiple ankyrin repeat domains protein 2, ... | | Authors: | Bento, I, Gracia Alai, M, Kreienkamp, J.-H. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Structural deficits in key domains of Shank2 lead to alterations in postsynaptic nanoclusters and to a neurodevelopmental disorder in humans.
Mol Psychiatry, 2022
|
|
7KEV
 
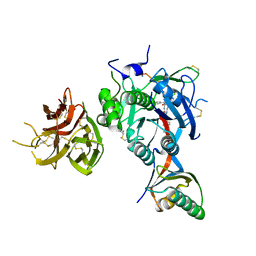 | | PCSK9 in complex with a cyclic peptide LDLR disruptor | | Descriptor: | CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, Proprotein convertase subtilisin/kexin type 9 Propeptide, ... | | Authors: | Spraggon, G, Chopra, R. | | Deposit date: | 2020-10-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
6QA1
 
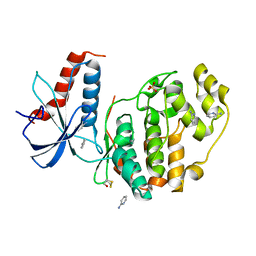 | | ERK2 mini-fragment binding | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, pyridin-2-amine | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
6FKU
 
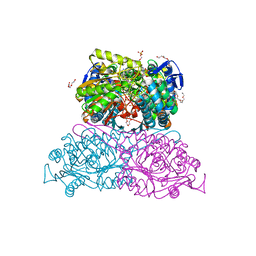 | | Structure and function of aldehyde dehydrogenase from Thermus thermophilus: An enzyme with an evolutionarily-distinct C-terminal arm (Recombinant protein with shortened C-terminal, in complex with NADP) | | Descriptor: | Aldehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hayes, K.A, Noor, M.R, Djeghader, A, Soulimane, T. | | Deposit date: | 2018-01-24 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The quaternary structure of Thermus thermophilus aldehyde dehydrogenase is stabilized by an evolutionary distinct C-terminal arm extension.
Sci Rep, 8, 2018
|
|
6D0K
 
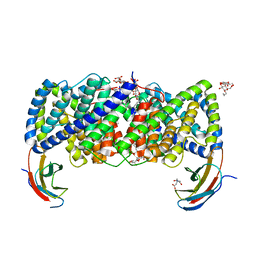 | | Crystal structure of a CLC-type fluoride/proton antiporter, E118Q mutant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7L7R
 
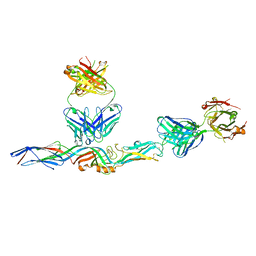 | |
6Q3P
 
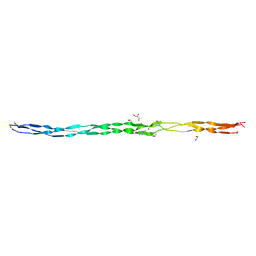 | | Atomic resolution crystal structure of an AAB collagen heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Leading Chain of the AAB Collagen Heterotrimer, ... | | Authors: | Jalan, A.A, Hartgerink, J.D, Brear, P, Leitinger, B, Farndale, R.W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution crystal structure of an AAB collagen heterotrimer
Nat.Chem.Biol., 2019
|
|
6FHB
 
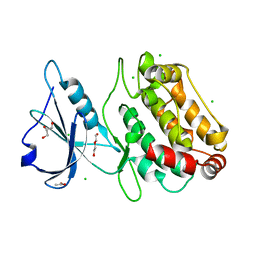 | |
6QA6
 
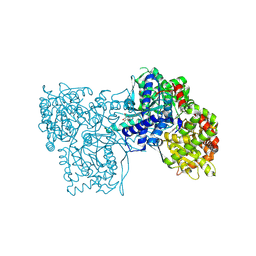 | | Glycogen Phosphorylase b in complex with 30 | | Descriptor: | (5~{S},7~{R},8~{S},9~{S},10~{R})-7-(hydroxymethyl)-2-naphthalen-2-yl-8,9,10-tris(oxidanyl)-6-oxa-1,3-diazaspiro[4.5]dec-1-en-4-one, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glucopyranosylidene-spiro-imidazolinones, a New Ring System: Synthesis and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetics and X-ray Crystallography.
J.Med.Chem., 62, 2019
|
|
7KH3
 
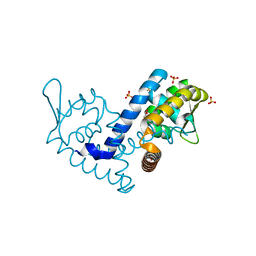 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole 3 propionic acid | | Descriptor: | INDOLYLPROPIONIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-19 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
6YPL
 
 | |
5H21
 
 | | Trimethoxy-ring inhibitor in complex with the first bromodomain of BRD4 | | Descriptor: | 3,4,5-trimethoxy-~{N}-(2-thiophen-2-ylethyl)benzamide, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Discovery of novel trimethoxy-ring BRD4 bromodomain inhibitors: AlphaScreen assay, crystallography and cell-based assay.
Medchemcomm, 8, 2017
|
|
