3QHD
 
 | |
2EUN
 
 | | Cytochrome c peroxidase (CCP) in complex with 2,4-diaminopyrimidine | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PYRIMIDINE-2,4-DIAMINE, cytochrome c peroxidase | | Authors: | Brenk, R, Vetter, S.W, Boyce, S.E, Goodin, D.B, Shoichet, B.K. | | Deposit date: | 2005-10-29 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing molecular docking in a charged model binding site.
J.Mol.Biol., 357, 2006
|
|
5VO0
 
 | | Structure of a TRAF6-Ubc13~Ub complex | | Descriptor: | POTASSIUM ION, TNF receptor-associated factor 6, Ubiquitin, ... | | Authors: | Middleton, A.J, Day, C.L. | | Deposit date: | 2017-05-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1.
Nat Commun, 8, 2017
|
|
5LOM
 
 | | Crystal structure of the PBP SocA from Agrobacterium tumefaciens C58 in complex with DFG at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Deoxyfructosyl-amino Acid Transporter Periplasmic Binding Protein, Deoxyfructosylglutamine | | Authors: | Marty, L, Vigouroux, A, Morera, S. | | Deposit date: | 2016-08-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for High Specificity of Amadori Compound and Mannopine Opine Binding in Bacterial Pathogens.
J.Biol.Chem., 291, 2016
|
|
5W1E
 
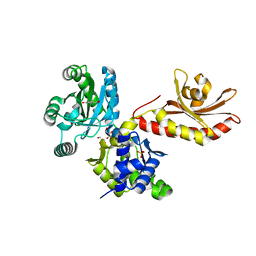 | | PobR in complex with PHB | | Descriptor: | GLYCEROL, P-HYDROXYBENZOIC ACID, Putative transcriptional regulator, ... | | Authors: | Page, R, Peti, W, Lord, D.M, Bajaj, R, Zhang, R. | | Deposit date: | 2017-06-02 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A peculiar IclR family transcription factor regulates para-hydroxybenzoate catabolism in Streptomyces coelicolor.
Nucleic Acids Res., 46, 2018
|
|
6NT5
 
 | | Cryo-EM structure of full-length human STING in the apo state | | Descriptor: | Stimulator of interferon protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
5VNZ
 
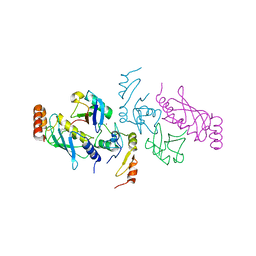 | | Structure of a TRAF6-Ubc13~Ub complex | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Middleton, A.J, Day, C.L. | | Deposit date: | 2017-05-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1.
Nat Commun, 8, 2017
|
|
2IJ5
 
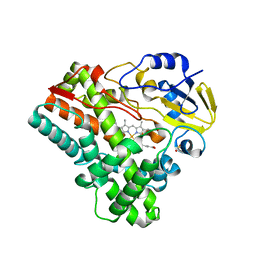 | |
5LOV
 
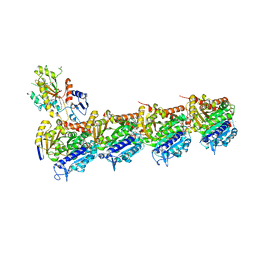 | | DZ-2384 tubulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DZ 2384, ... | | Authors: | Prota, A.E, Steinmetz, M.O, Shore, G.C, Brouhard, G, Roulston, A. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The synthetic diazonamide DZ-2384 has distinct effects on microtubule curvature and dynamics without neurotoxicity.
Sci Transl Med, 8, 2016
|
|
6H22
 
 | | Crystal structure of Mdm2 bound to a stapled peptide | | Descriptor: | 12-(dimethylamino)-3,10-diethyl-N,N,N-trimethyl-3,10-dihydrodibenzo[3,4:7,8]cycloocta[1,2-d:5,6-d']bis([1,2,3]triazole)-5-aminium, E3 ubiquitin-protein ligase Mdm2, Stapled peptide | | Authors: | Wang, X, Sharma, K, Spring, D.R, Hyvonen, M. | | Deposit date: | 2018-07-12 | | Release date: | 2019-07-31 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Water-soluble, stable and azide-reactive strained dialkynes for biocompatible double strain-promoted click chemistry.
Org.Biomol.Chem., 17, 2019
|
|
3N10
 
 | | Product complex of adenylate cyclase class IV | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Adenylate cyclase 2, MANGANESE (II) ION, ... | | Authors: | Gallagher, D.T, Reddy, P.T. | | Deposit date: | 2010-05-14 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active-Site Structure of Class IV Adenylyl Cyclase and Transphyletic Mechanism.
J.Mol.Biol., 405, 2011
|
|
4LTW
 
 | | Ancestral Ketosteroid Receptor-Progesterone-Mifepristone Complex | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, Ancestral Steroid Receptor 2, GLYCEROL, ... | | Authors: | Ortlund, E.A, Colucci, J.K. | | Deposit date: | 2013-07-24 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | X-ray crystal structure of the ancestral 3-ketosteroid receptor-progesterone-mifepristone complex shows mifepristone bound at the coactivator binding interface.
Plos One, 8, 2013
|
|
5VVW
 
 | |
7QWF
 
 | | BAZ2A bromodomain in complex with acetylpyrrole derivative compound 45 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-ethyl-2-methyl-5-[2-[2-(methylamino)ethyl]-1,3-thiazol-4-yl]-1~{H}-pyrrol-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2A, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-01-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
3NB6
 
 | |
6TLI
 
 | | THERMOLYSIN (60% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-29 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
5EL2
 
 | |
5MNA
 
 | | Cationic trypsin in complex with aniline (deuterated sample at 295 K) | | Descriptor: | ANILINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Charges Shift Protonation: Neutron Diffraction Reveals that Aniline and 2-Aminopyridine Become Protonated Upon Binding to Trypsin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MON
 
 | | Joint X-ray/neutron structure of cationic trypsin in complex with 2-aminopyridine | | Descriptor: | 2-AMINOPYRIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Schrader, T.E, Ostermann, A, Heine, A, Klebe, G. | | Deposit date: | 2016-12-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (0.939 Å), X-RAY DIFFRACTION | | Cite: | Charges Shift Protonation: Neutron Diffraction Reveals that Aniline and 2-Aminopyridine Become Protonated Upon Binding to Trypsin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4CVL
 
 | | PaMurF in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | Authors: | Ruane, K.M, Majce, V. | | Deposit date: | 2014-03-28 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Pamurf in Complex with AMP-Pnp
To be Published
|
|
3NB7
 
 | |
5MN1
 
 | | Cationic trypsin in complex with 2-aminopyridine (deuterated sample at 100 K) | | Descriptor: | 2-AMINOPYRIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.79 Å) | | Cite: | Charges Shift Protonation: Neutron Diffraction Reveals that Aniline and 2-Aminopyridine Become Protonated Upon Binding to Trypsin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MNY
 
 | | Neutron structure of cationic trypsin in complex with aniline | | Descriptor: | CALCIUM ION, Cationic trypsin, phenylazanium | | Authors: | Schiebel, J, Schrader, T.E, Ostermann, A, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | NEUTRON DIFFRACTION (1.43 Å) | | Cite: | Charges Shift Protonation: Neutron Diffraction Reveals that Aniline and 2-Aminopyridine Become Protonated Upon Binding to Trypsin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MNB
 
 | | Cationic trypsin in complex with 2-aminopyridine (deuterated sample at 295 K) | | Descriptor: | 2-AMINOPYRIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.939 Å) | | Cite: | Charges Shift Protonation: Neutron Diffraction Reveals that Aniline and 2-Aminopyridine Become Protonated Upon Binding to Trypsin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MNC
 
 | | Cationic trypsin in complex with aniline (deuterated sample at 100 K) | | Descriptor: | ANILINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.916 Å) | | Cite: | Charges Shift Protonation: Neutron Diffraction Reveals that Aniline and 2-Aminopyridine Become Protonated Upon Binding to Trypsin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
