4I6R
 
 | |
6V59
 
 | |
6EU9
 
 | | Crystal structure of Platynereis dumerilii RAR ligand-binding domain in complex with all-trans retinoic acid | | Descriptor: | RETINOIC ACID, Retinoic acid receptor | | Authors: | Handberg-Thorsager, M, Gutierrez-Mazariegos, J, Arold, S.T, Nadendla, E.K, Bertucci, P.Y, Germain, P, Tomancak, P, Pierzchalski, K, Jones, J.W, Albalat, R, Kane, M.A, Bourguet, W, Laudet, V, Arendt, D, Schubert, M. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The ancestral retinoic acid receptor was a low-affinity sensor triggering neuronal differentiation.
Sci Adv, 4, 2018
|
|
5BRD
 
 | | Crystal structure of Trypanosoma cruzi glucokinase in complex with inhibitor BENZ-GlcN | | Descriptor: | 2-(benzoylamino)-2-deoxy-beta-D-glucopyranose, Glucokinase 1, putative | | Authors: | D'Antonio, E.L, Perry, K, Deinema, M.S, Kearns, S.P, Frey, T.A. | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based approach to the identification of a novel group of selective glucosamine analogue inhibitors of Trypanosoma cruzi glucokinase.
Mol.Biochem.Parasitol., 204, 2016
|
|
8D4J
 
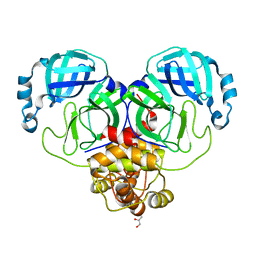 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
6URY
 
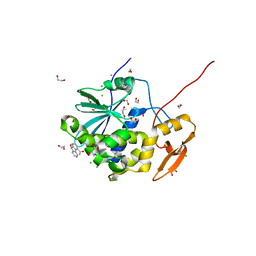 | | Crystal structure of ricin A chain in complex with inhibitor 9-oxo-4-fluorenecarboxamide | | Descriptor: | 1,2-ETHANEDIOL, 9-oxo-9H-fluorene-4-carboxamide, CHLORIDE ION, ... | | Authors: | Harijan, R.K, Li, X.P, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Small Molecule Inhibitors Targeting the Interaction of Ricin Toxin A Subunit with Ribosomes.
Acs Infect Dis., 6, 2020
|
|
6EWY
 
 | | RipA Peptidoglycan hydrolase (Rv1477, Mycobacterium tuberculosis) N-terminal domain | | Descriptor: | Peptidoglycan endopeptidase RipA | | Authors: | Schnell, R, Steiner, E.M, Schneider, G, Guy, J, Bourenkov, G. | | Deposit date: | 2017-11-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the N-terminal module of the cell wall hydrolase RipA and its role in regulating catalytic activity.
Proteins, 86, 2018
|
|
1S95
 
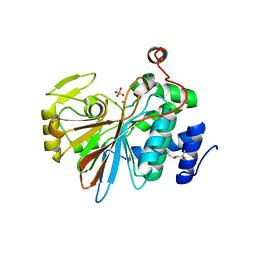 | | Structure of serine/threonine protein phosphatase 5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Swingle, M.R, Honkanen, R.E, Ciszak, E.M. | | Deposit date: | 2004-02-03 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the catalytic activity of human serine/threonine protein phosphatase-5.
J.Biol.Chem., 279, 2004
|
|
6F2S
 
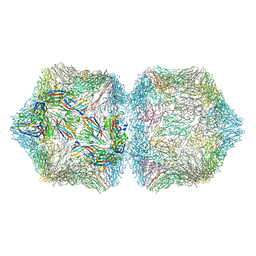 | | CryoEM structure of Ageratum Yellow Vein virus (AYVV) | | Descriptor: | Capsid protein, coat protein subunit H, coat protein subunit I, ... | | Authors: | Hesketh, E.L, Saunders, K, Fisher, C, Potze, J, Stanley, J, Lomonossoff, G.P, Ranson, N.A. | | Deposit date: | 2017-11-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The 3.3 angstrom structure of a plant geminivirus using cryo-EM.
Nat Commun, 9, 2018
|
|
4HTJ
 
 | |
3FPX
 
 | | Native fungus laccase from Trametes hirsuta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Stepanova, E.V, Cherkashin, E.A, Kurzeev, S.A, Strokopytov, B.V, Lamzin, V.S, Koroleva, O.V. | | Deposit date: | 2009-01-06 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of native laccase from Coriolus hirsutus at 1.8 A resolution
To be Published
|
|
1DWV
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and 4-amino tetrahydrobiopterin | | Descriptor: | 2,4-DIAMINO-6-[2,3-DIHYDROXY-PROP-3-YL]-5,6,7,8-TETRAHYDROPTERIDINE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
3NXP
 
 | | Crystal structure of human prethrombin-1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Chen, Z, Bush-Pelc, L.A, Di Cera, E. | | Deposit date: | 2010-07-14 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of prethrombin-1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6URX
 
 | | Crystal structure of ricin A chain in complex with inhibitor 5-phenyl-2-thiophenecarboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-phenylthiophene-2-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Harijan, R.K, Li, X.P, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Small Molecule Inhibitors Targeting the Interaction of Ricin Toxin A Subunit with Ribosomes.
Acs Infect Dis., 6, 2020
|
|
6V16
 
 | | Crystal structure of the bromodomain of human BRD7 bound to TP472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 7, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
1DST
 
 | |
7T5U
 
 | | Structure of E. coli MS115-1 CapH N-terminal domain | | Descriptor: | Helix-turn-helix domain-containing protein, SULFATE ION | | Authors: | Lau, R.K, Corbett, K.D. | | Deposit date: | 2021-12-13 | | Release date: | 2022-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | A conserved signaling pathway activates bacterial CBASS immune signaling in response to DNA damage.
Embo J., 41, 2022
|
|
7T5V
 
 | |
5B1A
 
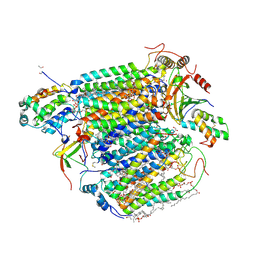 | | Bovine heart cytochrome c oxidase in the fully oxidized state at 1.5 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Yano, N, Muramoto, K, Shimada, A, Takemura, S, Baba, J, Fujisawa, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2015-12-01 | | Release date: | 2016-09-14 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Mg2+-containing Water Cluster of Mammalian Cytochrome c Oxidase Collects Four Pumping Proton Equivalents in Each Catalytic Cycle.
J.Biol.Chem., 291, 2016
|
|
6V4C
 
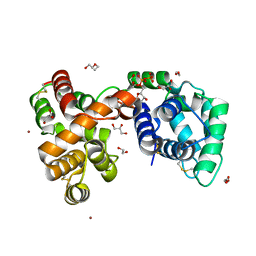 | | Culex quinquefasciatus D7 long form 1- CxD7L1 in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Calvo, E, Garboczi, D.N, Martin-Martin, I, Gittis, A.G. | | Deposit date: | 2019-11-27 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | ADP binding by the Culex quinquefasciatus mosquito D7 salivary protein enhances blood feeding on mammals.
Nat Commun, 11, 2020
|
|
3EZZ
 
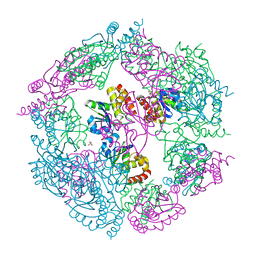 | | Crystal Structure of human MKP-2 | | Descriptor: | Dual specificity protein phosphatase 4, SULFATE ION | | Authors: | Jeong, D.G, Jung, S.K, Ryu, S.E, Kim, S.J. | | Deposit date: | 2008-10-24 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the catalytic domain of human MKP-2 reveals a 24-mer assembly.
Proteins, 76, 2009
|
|
7AKH
 
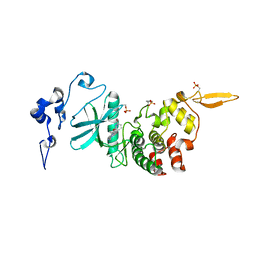 | | Structure of DYRK2 in complex with compound 58 | | Descriptor: | 4-[3-[(2~{S})-2-(6-bromanylpyridin-2-yl)oxypropyl]-2-methyl-imidazo[4,5-b]pyridin-5-yl]pyridine-2,6-diamine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-10-01 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AKL
 
 | | Structure of DYRK1A in complex with compound 50 | | Descriptor: | 4-(2,3-dimethyl-1-benzofuran-5-yl)pyridine-2,6-diamine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-10-01 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AKF
 
 | | Structure of DYRK2 in complex with compound 50 | | Descriptor: | 4-(2,3-dimethyl-1-benzofuran-5-yl)pyridine-2,6-diamine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
5TM2
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 2,5-bis(2-chloro-4-hydroxyphenyl)thiophene 1-oxide | | Descriptor: | 2,5-bis(2-chloro-4-hydroxyphenyl)-1H-1lambda~4~-thiophen-1-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
