3MMN
 
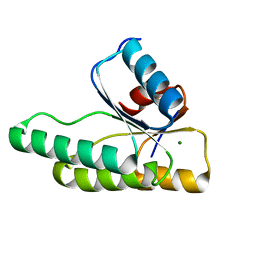 | | Crystal structure of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana complexed with Mg2+ | | Descriptor: | Histidine kinase homolog, MAGNESIUM ION | | Authors: | Marek, J, Klumpler, T, Pekarova, B, Triskova, O, Horak, J, Zidek, L, Dopitova, R, Hejatko, J, Janda, L. | | Deposit date: | 2010-04-20 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and binding specificity of the receiver domain of sensor histidine kinase CKI1 from Arabidopsis thaliana.
Plant J., 67, 2011
|
|
6Y13
 
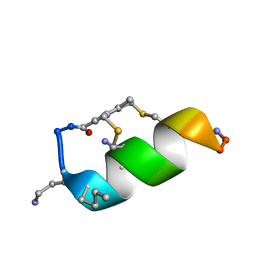 | |
6F2G
 
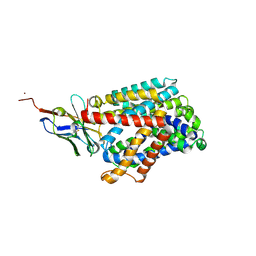 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | Nanobody 74, Putative amino acid/polyamine transport protein, ZINC ION | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
3F6I
 
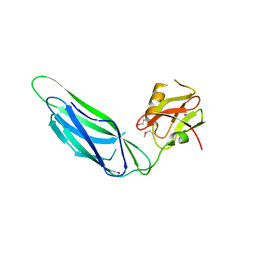 | | Structure of the SeMet labeled F4 fibrial chaperone FaeE | | Descriptor: | Chaperone protein faeE | | Authors: | Van Molle, I, Moonens, K, Buts, L, Garcia-Pino, A, Wyns, L, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | The F4 fimbrial chaperone FaeE is stable as a monomer that does not require self-capping of its pilin-interactive surfaces
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6F2W
 
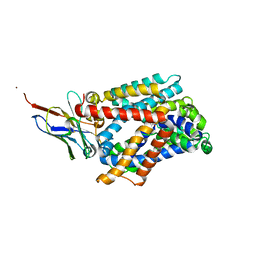 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | ALPHA-AMINOISOBUTYRIC ACID, Nanobody 74, Putative amino acid/polyamine transport protein, ... | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
3FFG
 
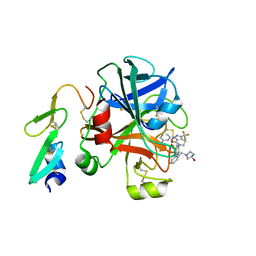 | | Factor XA in complex with the inhibitor (R)-6-(2'-((3- HYDROXYPYRROLIDIN-1-YL)METHYL)BIPHENYL-4-YL)-1-(3-(5-OXO-4,5-DIHYDRO-1H-1,2,4-TRIAZOL-3-YL)PHENYL)-3-(TRIFLUOROMETHYL)-5,6-DIHYDRO-1H-PYRAZOLO[3,4-C]PYRIDIN- 7(4H)-ONE | | Descriptor: | (R)-6-(2'-((3-hydroxypyrrolidin-1-yl)methyl)biphenyl-4-yl)-1-(3-(5-oxo-4,5-dihydro-1h-1,2,4-triazol-3-yl)phenyl)-3-(trifluoromethyl)-5,6-dihydro-1h-pyrazolo[3,4-c]pyridin-7(4h)-one, Coagulation factor X, heavy chain, ... | | Authors: | Alexander, R.A. | | Deposit date: | 2008-12-03 | | Release date: | 2009-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Phenyltriazolinones as potent factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6YF1
 
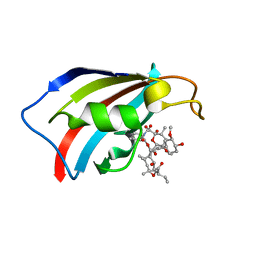 | | FKBP12 in complex with the BMP potentiator compound 8 at 1.12A resolution | | Descriptor: | (1aR,3R,5S,6R,7S,9R,10R,17aS,20S,21R,22S,25R,25aR)-25-Ethyl-10,22-dihydroxy-20-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-5,7-dimethoxy-1a,3,9,21-tetramethyloctadecahydro-2H-6,10-epoxyoxireno[p]pyrido[2,1-c][1,4]oxazacyclotricosine-11,12,18,24(1aH,14H)-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6YF2
 
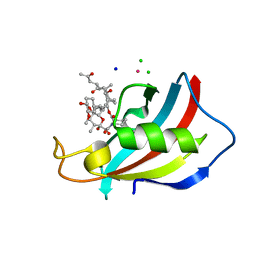 | | FKBP12 in complex with the BMP potentiator compound 6 at 1.03A resolution | | Descriptor: | (1~{R},9~{S},12~{S},13~{R},14~{S},17~{R},18~{E},21~{S},23~{S},24~{R},25~{S},27~{R})-23,25-dimethoxy-12-[(~{E})-1-[(1~{R},3~{R},4~{R})-3-methoxy-4-oxidanyl-cyclohexyl]prop-1-en-2-yl]-13,19,21,27-tetramethyl-1,14-bis(oxidanyl)-17-(2-oxidanylidenepropyl)-11,28-dioxa-4-azatricyclo[22.3.1.0^{4,9}]octacos-18-ene-2,3,10,16-tetrone, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6YF0
 
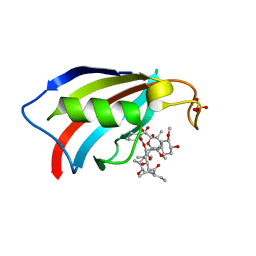 | | FKBP12 in complex with the BMP potentiator compound 9 at 1.55 A resolution | | Descriptor: | 18-HYDROXYASCOMYCIN, Peptidyl-prolyl cis-trans isomerase FKBP1A, SULFATE ION | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6YF3
 
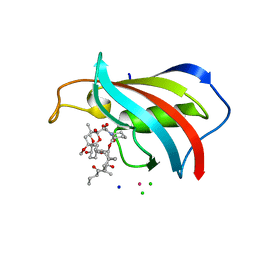 | | FKBP12 in complex with the BMP potentiator compound 10 at 1.00A resolution | | Descriptor: | (1~{R},9~{S},12~{S},13~{R},14~{S},17~{R},18~{E},21~{S},23~{S},24~{R},25~{S},27~{R})-17-ethyl-25-methoxy-12-[(~{E})-1-[(1~{R},3~{R},4~{R})-3-methoxy-4-oxidanyl-cyclohexyl]prop-1-en-2-yl]-13,19,21,27-tetramethyl-1,14,23-tris(oxidanyl)-11,28-dioxa-4-azatricyclo[22.3.1.0^{4,9}]octacos-18-ene-2,3,10,16-tetrone, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
3M6F
 
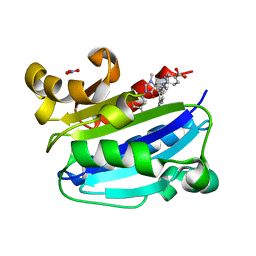 | | CD11A I-domain complexed with 6-((5S,9R)-9-(4-CYANOPHENYL)-3-(3,5-DICHLOROPHENYL)-1-METHYL-2,4-DIOXO-1,3,7- TRIAZASPIRO[4.4]NON-7-YL)NICOTINIC ACID | | Descriptor: | 6-[(5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]non-7-yl]pyridine-3-carboxylic acid, Integrin alpha-L, NITRATE ION | | Authors: | Sheriff, S. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small molecule antagonist of leukocyte function associated antigen-1 (LFA-1): structure-activity relationships leading to the identification of 6-((5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]nonan-7-yl)nicotinic acid (BMS-688521).
J.Med.Chem., 53, 2010
|
|
3D4Q
 
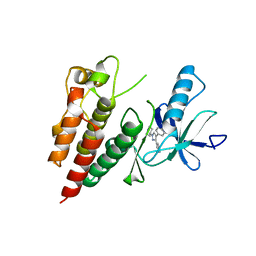 | | Pyrazole-based inhibitors of B-Raf kinase | | Descriptor: | (1E)-5-(1-piperidin-4-yl-3-pyridin-4-yl-1H-pyrazol-4-yl)-2,3-dihydro-1H-inden-1-one oxime, B-Raf proto-oncogene serine/threonine-protein kinase | | Authors: | Morales, T, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2008-05-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent and selective pyrazole-based inhibitors of B-Raf kinase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3MET
 
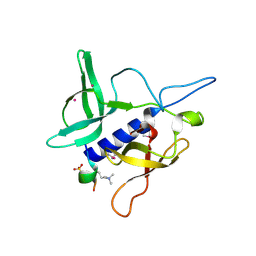 | | Crystal structure of SGF29 in complex with H3K4me2 | | Descriptor: | GLYCEROL, Histone H3, SAGA-associated factor 29 homolog, ... | | Authors: | Bian, C.B, Xu, C, Lam, R, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
3D23
 
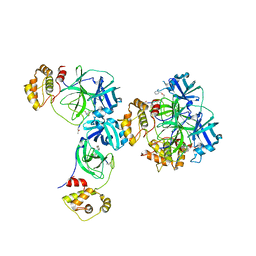 | | Main protease of HCoV-HKU1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhao, Q, Chen, C, Li, S, Zou, Y. | | Deposit date: | 2008-05-07 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1.
J.Virol., 82, 2008
|
|
6PL9
 
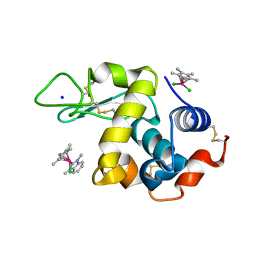 | | Adduct formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimidaz ol-2-ylidene)(eta5-pentamethylcyclopentadienyl)rhodium(III) with HEWL | | Descriptor: | 2-(1-chloranyl-2,3,4,5,6-pentamethyl-1$l^{7}-rhodapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexan-1-yl)-1,3-dimethyl-benzimidazole, Lysozyme, SODIUM ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Probing the Paradigm of Promiscuity for N-Heterocyclic Carbene Complexes and their Protein Adduct Formation.
Angew.Chem.Int.Ed.Engl., 2021
|
|
3D38
 
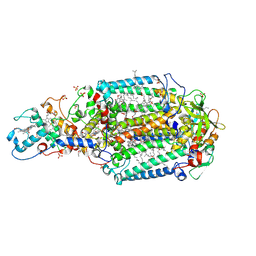 | | Crystal structure of new trigonal form of photosynthetic reaction center from Blastochloris viridis. Crystals grown in microfluidics by detergent capture. | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Li, L, Nachtergaele, S.H.M, Seddon, A.M, Tereshko, V, Ponomarenko, N, Ismagilov, R.F, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Simple host-guest chemistry to modulate the process of concentration and crystallization of membrane proteins by detergent capture in a microfluidic device.
J.Am.Chem.Soc., 130, 2008
|
|
3LDS
 
 | | Crystal structure of RB69 gp43 with DNA and dATP opposite 8-oxoG | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*(8OG)P*CP*TP*TP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*AP*AP*(DDG))-3'), ... | | Authors: | Hogg, M, Midkiff, J, Rudnicki, J, Reha-Krantz, L, Doublie, S, Wallace, S.S. | | Deposit date: | 2010-01-13 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinetics of mismatch formation opposite lesions by the replicative DNA polymerase from bacteriophage RB69.
Biochemistry, 49, 2010
|
|
6PQ9
 
 | | Crystal Structure of TLA-1 S70G extended spectrum Beta-lactamase | | Descriptor: | ACETATE ION, ASPARTIC ACID, Beta-lactamase, ... | | Authors: | Rudino-Pinera, E, Cifuentes-Castro, V.H, Rodriguez-Alamazan, C. | | Deposit date: | 2019-07-08 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | The crystal structure of ESBL TLA-1 in complex with clavulanic acid reveals a second acylation site.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
6YDZ
 
 | | Human wtSTING in complex with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3CN0
 
 | |
3LMC
 
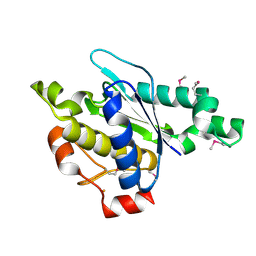 | | Crystal Structure of zinc-dependent peptidase from Methanocorpusculum labreanum (strain Z), Northeast Structural Genomics Consortium Target MuR16 | | Descriptor: | FE (III) ION, Peptidase, zinc-dependent, ... | | Authors: | Kuzin, A, Ashok, S, Vorobiev, S, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Northeast Structural Genomics Consortium Target MuR16
To be published
|
|
3LML
 
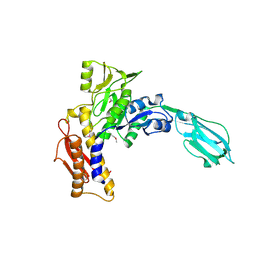 | | Crystal Structure of the sheath tail protein Lin1278 from Listeria innocua, Northeast Structural Genomics Consortium Target LkR115 | | Descriptor: | Lin1278 protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Patel, D.J, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target LkR115
To be Published
|
|
3LOE
 
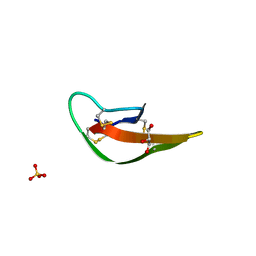 | |
6YEA
 
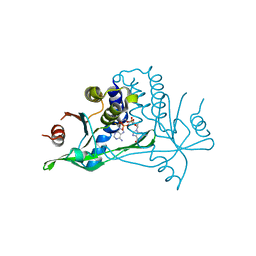 | | Human wtSTING in complex with 2',2'-difluoro-3',3'-cGAMP | | Descriptor: | 2',2'-difluoro-3',3'-cGAMP, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3LO1
 
 | |
