3BDJ
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with a Covalently Bound Oxipurinol Inhibitor | | Descriptor: | CALCIUM ION, CARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F, Nishino, T. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of inhibition of xanthine oxidoreductase by allopurinol: crystal structure of reduced bovine milk xanthine oxidoreductase bound with oxipurinol.
Nucleosides Nucleotides Nucleic Acids, 27, 2008
|
|
5XPP
 
 | | Crystal structure of VDR-LBD complexed with 25RS-(Hydroxyphenyl)-2-methylidene-19,26,27-trinor-1,25-dihydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-oxidanyl-hexan-2-yl]-7~{ a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
4H95
 
 | |
3RN1
 
 | |
6B3D
 
 | |
3RMZ
 
 | |
1JQA
 
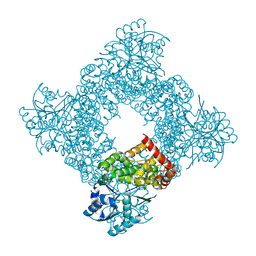 | | Bacillus stearothermophilus glycerol dehydrogenase complex with glycerol | | Descriptor: | GLYCEROL, Glycerol Dehydrogenase, ZINC ION | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-04 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
2I7S
 
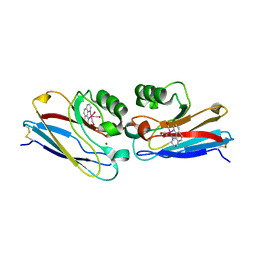 | | Crystal structure of Re(phen)(CO)3 (Thr124His)(His83Gln) Azurin Cu(II) from Pseudomonas aeruginosa | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COBALT TETRAAMMINE ION, ... | | Authors: | Gradinaru, C, Crane, B.R. | | Deposit date: | 2006-08-31 | | Release date: | 2006-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Relaxation dynamics of Pseudomonas aeruginosa Re(I)(CO)3(alpha-diimine)(HisX)+ (X = 83, 107, 109, 124, 126)Cu(II) azurins.
J.Am.Chem.Soc., 131, 2009
|
|
4Y9Z
 
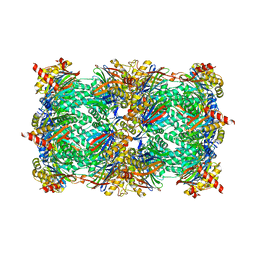 | |
4D8G
 
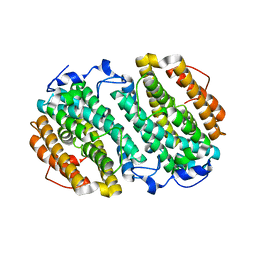 | | Chlamydia trachomatis NrdB with a Mn/Fe cofactor (procedure 2 - low Mn) | | Descriptor: | FE (III) ION, MANGANESE (II) ION, Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Dassama, L.M.K, Boal, A.K, Krebs, C, Rosenzweig, A.C, Bollinger Jr, J.M. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Evidence that the beta subunit of Chlamydia trachomatis ribonucleotide reductase is active with the manganese ion of its manganese(IV)/iron(III) cofactor in site 1.
J.Am.Chem.Soc., 134, 2012
|
|
3FLK
 
 | | Crystal Structure of Tartrate Dehydrogenase from Pseudomonas putida in complex with NADH, oxalate and metal ion | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, AMMONIUM ION, ... | | Authors: | Malik, R, Viola, R.E. | | Deposit date: | 2008-12-18 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of tartrate dehydrogenase: a versatile enzyme catalyzing multiple reactions.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7KXA
 
 | |
5U3J
 
 | | Crystal Structure of DH511.1 Fab in Complex with HIV-1 gp41 MPER Peptide | | Descriptor: | DH511.1 Heavy Chain, DH511.1 Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
3F0Q
 
 | | Staphylococcus aureus dihydrofolate reductase complexed with NADPH and 2,4-Diamino-5-[3-(3-methoxy-5-(2,6-dimethylphenyl)phenyl)but-1-ynyl]-6-methylpyrimidine | | Descriptor: | 5-[(3S)-3-(5-methoxy-2',6'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2008-10-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Predicting resistance mutations using protein design algorithms.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4KNE
 
 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
3NVW
 
 | | Crystal Structure of Bovine Xanthine Oxidase in Complex with Guanine | | Descriptor: | DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cao, H, Hille, R. | | Deposit date: | 2010-07-08 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate orientation and specificity in xanthine oxidase: crystal structures of the enzyme in complex with indole-3-acetaldehyde and guanine.
Biochemistry, 53, 2014
|
|
5XPM
 
 | | Crystal structure of VDR-LBD complexed with 22S-Butyl-25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},3~{S})-3-[(3~{S})-3-(4-hydroxyphenyl)-3-methoxy-propyl]heptan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
1BPW
 
 | | BETAINE ALDEHYDE DEHYDROGENASE FROM COD LIVER | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (ALDEHYDE DEHYDROGENASE) | | Authors: | Johansson, K, El Ahmad, M, Ramaswamy, S, Hjelmqvist, L, Jornvall, H, Eklund, H. | | Deposit date: | 1998-08-12 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of betaine aldehyde dehydrogenase at 2.1 A resolution.
Protein Sci., 7, 1998
|
|
7LJU
 
 | | Porcine Dihydropyrimidine Dehydrogenase (DPD) crosslinked with 5-Ethynyluracil (5EU) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 5-ethynylpyrimidine-2,4(1H,3H)-dione, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Butrin, A, Forouzesh, D, Beaupre, B, Wawrzak, Z, Liu, D, Moran, G. | | Deposit date: | 2021-01-30 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Interaction of Porcine Dihydropyrimidine Dehydrogenase with the Chemotherapy Sensitizer: 5-Ethynyluracil.
Biochemistry, 60, 2021
|
|
3BV7
 
 | | Crystal structure of Delta(4)-3-ketosteroid 5-beta-reductase in complex with NADP and glycerol. Resolution: 1.79 A. | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Di Costanzo, L, Drury, J, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-01-04 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of Human Liver {Delta}4-3-Ketosteroid 5{beta}-Reductase (AKR1D1) and Implications for Substrate Binding and Catalysis.
J.Biol.Chem., 283, 2008
|
|
2GS5
 
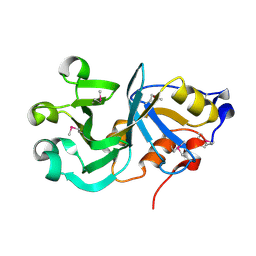 | |
3WZE
 
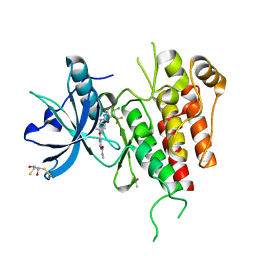 | | KDR in complex with ligand sorafenib | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, ACETATE ION, ... | | Authors: | Okamoto, K, Ikemori_Kawada, M, Inoue, A, Matsui, J. | | Deposit date: | 2014-09-24 | | Release date: | 2015-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct binding mode of multikinase inhibitor lenvatinib revealed by biochemical characterization.
ACS MED.CHEM.LETT., 6, 2015
|
|
1SXA
 
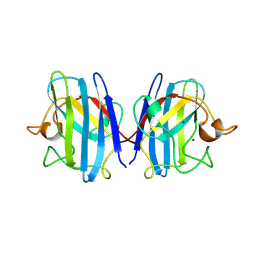 | | CRYSTAL STRUCTURE OF REDUCED BOVINE ERYTHROCYTE SUPEROXIDE DISMUTASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Rypniewski, W.R, Mangani, S, Bruni, B, Orioli, P, Casati, M, Wilson, K.S. | | Deposit date: | 1995-03-17 | | Release date: | 1995-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of reduced bovine erythrocyte superoxide dismutase at 1.9 A resolution.
J.Mol.Biol., 251, 1995
|
|
5EGE
 
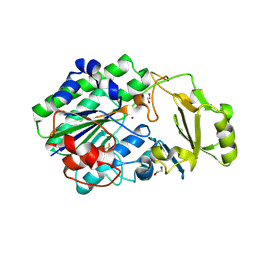 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
4GKI
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NM-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
