1XTY
 
 | | Crystal structure of Sulfolobus solfataricus peptidyl-tRNA hydrolase | | Descriptor: | Peptidyl-tRNA hydrolase, SULFATE ION | | Authors: | Fromant, M, Schmitt, E, Mechulam, Y, Lazennec, C, Plateau, P, Blanquet, S. | | Deposit date: | 2004-10-25 | | Release date: | 2005-03-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure at 1.8 A resolution and identification of active site residues of Sulfolobus solfataricus peptidyl-tRNA hydrolase.
Biochemistry, 44, 2005
|
|
1F5V
 
 | | STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF A FLAVOPROTEIN FROM ESCHERICHIA COLI THAT REDUCES NITROCOMPOUNDS. ALTERATION OF PYRIDINE NUCLEOTIDE BINDING BY A SINGLE AMINO ACID SUBSTITUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NADPH NITROREDUCTASE | | Authors: | Kobori, T, Sasaki, H, Lee, W.C, Zenno, S, Saigo, K, Murphy, M.E.P, Tanokura, M. | | Deposit date: | 2000-06-17 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution.
J.Biol.Chem., 276, 2001
|
|
4M3Q
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1917 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Zhang, W, Zhang, D, Stashko, M.A, DeRyckere, D, Hunter, D, Kireev, D.B, Miley, M, Cummings, C, Lee, M, Norris-Drouin, J, Stewart, W.M, Sather, S, Zhou, Y, Kirkpatrick, G, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Pseudo-Cyclization through Intramolecular Hydrogen Bond Enables Discovery of Pyridine Substituted Pyrimidines as New Mer Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
1Z4L
 
 | | Structure of the D41N variant of the human mitochondrial deoxyribonucleotidase in complex with thymidine 5'-monophosphate | | Descriptor: | 5'(3')-deoxyribonucleotidase, MAGNESIUM ION, THYMIDINE-5'-PHOSPHATE | | Authors: | Wallden, K, Ruzzenente, B, Rinaldo-Matthis, A, Bianchi, V, Nordlund, P. | | Deposit date: | 2005-03-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for substrate specificity of the human mitochondrial deoxyribonucleotidase
STRUCTURE, 13, 2005
|
|
1ZAW
 
 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P212121, Form A | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
1ZN2
 
 | | Low Resolution Structure of Response Regulator StyR | | Descriptor: | MAGNESIUM ION, response regulatory protein | | Authors: | Milani, M, Leoni, L, Rampioni, G, Zennaro, E, Ascenzi, P, Bolognesi, M. | | Deposit date: | 2005-05-11 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | An Active-like Structure in the Unphosphorylated StyR Response Regulator Suggests a Phosphorylation- Dependent Allosteric Activation Mechanism.
STRUCTURE, 13, 2005
|
|
1Z4K
 
 | | Structure of the D41N variant of the human mitochondrial deoxyribonucleotidase in complex with thymidine 3'-monophosphate | | Descriptor: | 5'(3')-deoxyribonucleotidase, MAGNESIUM ION, THYMIDINE-3'-PHOSPHATE | | Authors: | Wallden, K, Ruzzenente, B, Rinaldo-Matthis, A, Bianchi, V, Nordlund, P. | | Deposit date: | 2005-03-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for substrate specificity of the human mitochondrial deoxyribonucleotidase
STRUCTURE, 13, 2005
|
|
1Z4P
 
 | | Structure of the D41N variant of the human mitochondrial deoxyribonucleotidase in complex with deoxyriboguanosine 5'-monophosphate | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, 5'(3')-deoxyribonucleotidase, MAGNESIUM ION | | Authors: | Wallden, K, Ruzzenente, B, Rinaldo-Matthis, A, Bianchi, V, Nordlund, P. | | Deposit date: | 2005-03-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate specificity of the human mitochondrial deoxyribonucleotidase
STRUCTURE, 13, 2005
|
|
1Z8A
 
 | | Human Aldose Reductase complexed with novel Sulfonyl-pyridazinone Inhibitor | | Descriptor: | 6-[(5-CHLORO-3-METHYL-1-BENZOFURAN-2-YL)SULFONYL]PYRIDAZIN-3(2H)-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Podjarny, A, Heine, A, Klebe, G. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution crystal structure of aldose reductase complexed with the novel sulfonyl-pyridazinone inhibitor exhibiting an alternative active site anchoring group.
J.Mol.Biol., 356, 2006
|
|
1Z89
 
 | | Human Aldose Reductase complexed with novel Sulfonyl-pyridazinone Inhibitor | | Descriptor: | 6-[(5-CHLORO-3-METHYL-1-BENZOFURAN-2-YL)SULFONYL]PYRIDAZIN-3(2H)-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Podjarny, A, Heine, A, Klebe, G. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | High-resolution crystal structure of aldose reductase complexed with the novel sulfonyl-pyridazinone inhibitor exhibiting an alternative active site anchoring group.
J.Mol.Biol., 356, 2006
|
|
1ZAV
 
 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P21 | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
1Z4M
 
 | | Structure of the D41N variant of the human mitochondrial deoxyribonucleotidase in complex with uridine 5'-monophosphate | | Descriptor: | 5'(3')-deoxyribonucleotidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wallden, K, Ruzzenente, B, Rinaldo-Matthis, A, Bianchi, V, Nordlund, P. | | Deposit date: | 2005-03-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for substrate specificity of the human mitochondrial deoxyribonucleotidase
STRUCTURE, 13, 2005
|
|
3JCS
 
 | | 2.8 Angstrom cryo-EM structure of the large ribosomal subunit from the eukaryotic parasite Leishmania | | Descriptor: | 26S alpha ribosomal RNA, 26S delta ribosomal RNA, 26S epsilon ribosomal RNA, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Matzov, D, Halfon, Y, Zackay, A, Rozenberg, H, Zimmerman, E, Bashan, A, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-20 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | 2.8- angstrom Cryo-EM Structure of the Large Ribosomal Subunit from the Eukaryotic Parasite Leishmania.
Cell Rep, 16, 2016
|
|
1Z6T
 
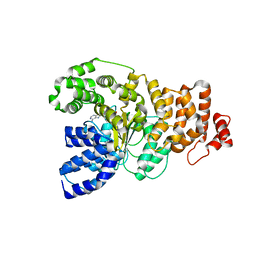 | | Structure of the apoptotic protease-activating factor 1 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Apoptotic protease activating factor 1 | | Authors: | Riedl, S.J, Li, W, Chao, Y, Schwarzenbacher, R, Shi, Y. | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the apoptotic protease-activating factor 1 bound to ADP
Nature, 434, 2005
|
|
1BKB
 
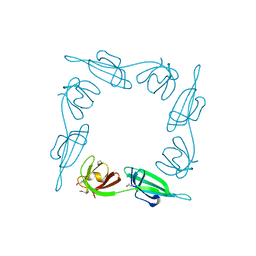 | | INITIATION FACTOR 5A FROM ARCHEBACTERIUM PYROBACULUM AEROPHILUM | | Descriptor: | TRANSLATION INITIATION FACTOR 5A | | Authors: | Peat, T.S, Newman, J, Waldo, G.S, Berendzen, J, Terwilliger, T.C. | | Deposit date: | 1998-07-05 | | Release date: | 1998-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 A resolution.
Structure, 6, 1998
|
|
2YOJ
 
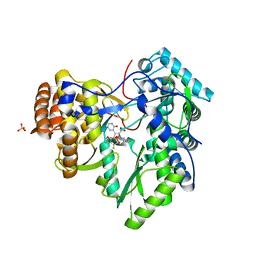 | | HCV NS5B polymerase complexed with pyridonylindole compound | | Descriptor: | 4-fluoranyl-6-[(7-fluoranyl-4-oxidanylidene-3H-quinazolin-6-yl)methyl]-8-(2-oxidanylidene-1H-pyridin-3-yl)furo[2,3-e]indole-7-carboxylic acid, PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Chen, K.X, Venkatraman, S, Anilkumar, G.N, Zeng, Q, Lesburg, C.A, Vibulbhan, B, Yang, W, Velazquez, F, Chan, T.-Y, Bennett, F, Sannigrahi, M, Jiang, Y, Duca, J.S, Pinto, P, Gavalas, S, Huang, Y, Wu, W, Selyutin, O, Agrawal, S, Feld, B, Huang, H.-C, Li, C, Cheng, K.-C, Shih, N.-Y, Kozlowski, J.A, Rosenblum, S.B, Njoroge, F.G. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of Sch 900188: A Potent Hepatitis C Virus Ns5B Polymerase Inhibitor Prodrug as a Development Candidate
Acs Med.Chem.Lett., 5, 2014
|
|
4E4U
 
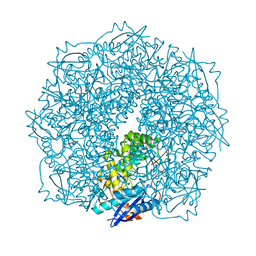 | | Crystal structure of a putative Mandelate racemase/Muconate lactonizing enzyme (Target PSI-200780) from Burkholderia SAR-1 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Mandalate racemase/muconate lactonizing enzyme | | Authors: | Kumar, P.R, Bonanno, J, Chowdhury, S, Foti, R, Gizzi, A, Hammonds, J, Hillerich, B, Matikainen, B, Seidel, R, Toro, R, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-13 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a putative MR/ML enzyme from Burkholderia SAR-1
to be published
|
|
4ECJ
 
 | | Crystal structure of glutathione s-transferase prk13972 (target efi-501853) from pseudomonas aeruginosa pacs2 complexed with glutathione | | Descriptor: | GLUTATHIONE, glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Prk13972 from Pseudomonas Aeruginosa
To be Published
|
|
3TWA
 
 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 complexed with magnesium and glycerol | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
2YFS
 
 | | Crystal structure of inulosucrase from Lactobacillus johnsonii NCC533 in complex with sucrose | | Descriptor: | CALCIUM ION, LEVANSUCRASE, SULFATE ION, ... | | Authors: | Pijning, T, Anwar, M.A, Leemhuis, H, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Inulosucrase from Lactobacillus: Insights Into the Substrate Specificity and Product Specificity of Gh68 Fructansucrases.
J.Mol.Biol., 412, 2011
|
|
2EX4
 
 | | Crystal Structure of Human methyltransferase AD-003 in complex with S-adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, adrenal gland protein AD-003 | | Authors: | Min, J.R, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-07 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of Human AD-003 protein in complex with S-adenosyl-L-homocysteine
To be Published
|
|
4EXJ
 
 | | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus | | Descriptor: | CHLORIDE ION, SULFATE ION, uncharacterized protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus
To be Published
|
|
2AQE
 
 | | Structural and functional analysis of ada2 alpha swirm domain | | Descriptor: | Transcriptional adaptor 2, Ada2 alpha | | Authors: | Qian, C, Zhang, Q, Zeng, L, Zhou, M.-M. | | Deposit date: | 2005-08-17 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and chromosomal DNA binding of the SWIRM domain
Nat.Struct.Mol.Biol., 12, 2005
|
|
5MCT
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LHG1) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
5TUA
 
 | | structure of a Na+-selective mutant of two-pore channel from Arabidopsis thaliana AtTPC1 | | Descriptor: | BARIUM ION, CALCIUM ION, SODIUM ION, ... | | Authors: | Guo, J, Zeng, W, Jiang, Y. | | Deposit date: | 2016-11-05 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tuning the ion selectivity of two-pore channels.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
