1KIG
 
 | | BOVINE FACTOR XA | | Descriptor: | ANTICOAGULANT PEPTIDE, FACTOR XA | | Authors: | Wei, A, Alexander, R, Chang, C.-H. | | Deposit date: | 1997-04-24 | | Release date: | 1998-10-28 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected binding mode of tick anticoagulant peptide complexed to bovine factor Xa.
J.Mol.Biol., 283, 1998
|
|
3BC1
 
 | | Crystal Structure of the complex Rab27a-Slp2a | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-27A, ... | | Authors: | Chavas, L.M.G, Ihara, K, Kawasaki, M, Wakatsuki, S. | | Deposit date: | 2007-11-12 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elucidation of Rab27 recruitment by its effectors: structure of Rab27a bound to Exophilin4/Slp2-a
Structure, 16, 2008
|
|
6GAA
 
 | | BACTERIORHODOPSIN, 430 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
1HPY
 
 | | THE SOLUTION STRUCTURE OF HUMAN PARATHYROID HORMONE FRAGMENT 1-34 IN 20% TRIFLUORETHANOL, NMR, 10 STRUCTURES | | Descriptor: | PARATHYROID HORMONE | | Authors: | Marx, U.C, Roesch, P, Adermann, K, Bayer, P, Forssmann, W.-G. | | Deposit date: | 1998-09-30 | | Release date: | 2000-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human parathyroid hormone fragments hPTH(1-34) and hPTH(1-39) and bovine parathyroid hormone fragment bPTH(1-37).
Biochem.Biophys.Res.Commun., 267, 2000
|
|
3BGB
 
 | | HIV-1 protease in complex with a isobutyl decorated oligoamine | | Descriptor: | CHLORIDE ION, N,N'-(iminodiethane-2,1-diyl)bis[4-amino-N-(2-methylpropyl)benzenesulfonamide], Protease | | Authors: | Boettcher, J, Blum, A, Sammet, B, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-11-26 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Achiral oligoamines as versatile tool for the development of aspartic protease inhibitors
Bioorg.Med.Chem., 16, 2008
|
|
1HRL
 
 | |
3PAL
 
 | | IONIC INTERACTIONS WITH PARVALBUMINS. CRYSTAL STRUCTURE DETERMINATION OF PIKE 4.10 PARVALBUMIN IN FOUR DIFFERENT IONIC ENVIRONMENTS | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J, Rambaud, J. | | Deposit date: | 1990-11-08 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ionic interactions with parvalbumins. Crystal structure determination of pike 4.10 parvalbumin in four different ionic environments.
J.Mol.Biol., 220, 1991
|
|
5VZG
 
 | | Pre-catalytic ternary complex of human Polymerase Mu (W434H) mutant with incoming nonhydrolyzable UMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, CHLORIDE ION, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
6GAE
 
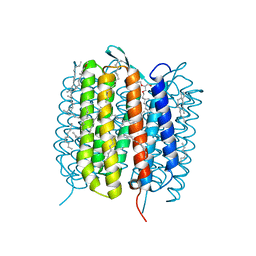 | | BACTERIORHODOPSIN, 560 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
5WM1
 
 | | Structure of the 10S (+)-trans-BP-dG modified Rev1 ternary complex | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Rechkoblit, O, Kolbanovsky, A, Landes, H, Geacintov, N.E, Aggarwal, A.K. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.
Nat Commun, 8, 2017
|
|
3DRU
 
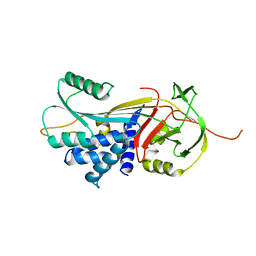 | | Crystal Structure of Gly117Phe Alpha1-Antitrypsin | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Gooptu, B, Nobeli, I, Purkiss, A, Phillips, R.L, Mallya, M, Lomas, D.A, Barrett, T.E. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic and cellular characterisation of two mechanisms stabilising the native fold of alpha1-antitrypsin: implications for disease and drug design.
J.Mol.Biol., 387, 2009
|
|
3TKG
 
 | | crystal structure of HIV model protease precursor/saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | Deposit date: | 2011-08-26 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
3RSX
 
 | |
3RTM
 
 | |
3BMP
 
 | |
3BNZ
 
 | | Crystal structure of Thymidylate Synthase ternary complex with dUMP and 8A inhibitor | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-(4-methyl-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)benzonitrile, PHOSPHATE ION, ... | | Authors: | Leone, R, Cancian, L, Luciani, R, Ferrari, S, Costi, M.P, Mangani, S. | | Deposit date: | 2007-12-15 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
3REC
 
 | | ESCHERICHIA COLI RECA PROTEIN-BOUND DNA, NMR, 1 STRUCTURE | | Descriptor: | DNA (5'-D(*TP*A)-3') | | Authors: | Nishinaka, T, Ito, Y, Yokoyama, S, Shibata, T. | | Deposit date: | 1997-04-17 | | Release date: | 1997-10-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An extended DNA structure through deoxyribose-base stacking induced by RecA protein.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
6GA8
 
 | | BACTERIORHODOPSIN, 330 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GAH
 
 | | BACTERIORHODOPSIN, 680 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
3RU1
 
 | |
5IP9
 
 | | Structure of RNA Polymerase II-TFIIF complex | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Transcription initiation complex structures elucidate DNA opening.
Nature, 533, 2016
|
|
3I5M
 
 | | Structure of the apo form of leucoanthocyanidin reductase from vitis vinifera | | Descriptor: | Putative leucoanthocyanidin reductase 1 | | Authors: | Mauge, C, Gargouri, M, d'Estaintot, B.L, Granier, T, Gallois, B. | | Deposit date: | 2009-07-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure and catalytic mechanism of leucoanthocyanidin reductase from Vitis vinifera.
J.Mol.Biol., 397, 2010
|
|
6G7J
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 457-646 fs state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
3FHZ
 
 | | Crystal structure of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator and co-repressor, L-arginine | | Descriptor: | 5'-D(*TP*GP*TP*TP*GP*CP*AP*TP*AP*AP*CP*GP*AP*TP*GP*CP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*CP*AP*TP*CP*GP*TP*TP*AP*TP*GP*CP*AP*AP*CP*A)-3', ACETATE ION, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | The structure of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator and Co-repressor, L-arginine.
J.Mol.Biol., 388, 2009
|
|
3BUH
 
 | | BACE-1 complexed with compound 4 | | Descriptor: | 4-(2-aminoethyl)-2-cyclohexylphenol, beta-secretase 1 | | Authors: | Kuglstatter, A, Hennig, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-03-11 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tyramine fragment binding to BACE-1
Bioorg.Med.Chem.Lett., 18, 2008
|
|
