5OM5
 
 | |
6K61
 
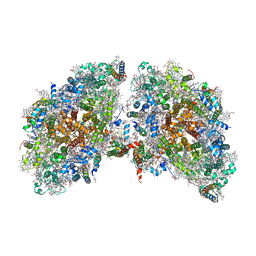 | | Cryo-EM structure of the tetrameric photosystem I from a heterocyst-forming cyanobacterium Anabaena sp. PCC7120 | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Zheng, L, Li, Y, Li, X, Zhong, Q, Li, N, Zhang, K, Zhang, Y, Chu, H, Ma, C, Li, G, Zhao, J, Gao, N. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural and functional insights into the tetrameric photosystem I from heterocyst-forming cyanobacteria.
Nat.Plants, 5, 2019
|
|
4J3I
 
 | | X-ray crystal structure of bromodomain complex to 1.24 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4, ... | | Authors: | Stein, A.J, White, A, Suto, R.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | RVX-208, an Inducer of ApoA-I in Humans, Is a BET Bromodomain Antagonist.
Plos One, 8, 2013
|
|
4J4H
 
 | | PylD in complex with pyrroline-carboxy-lysine and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, N~6~-[(2R)-3,4-dihydro-2H-pyrrol-2-ylcarbonyl]-L-lysine, ... | | Authors: | Quitterer, F, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2013-02-06 | | Release date: | 2013-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Reaction Mechanism of Pyrrolysine Synthase (PylD).
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
5XH7
 
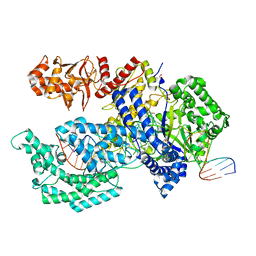 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RR variant in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
4J1N
 
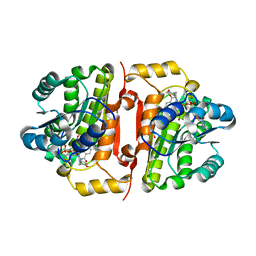 | | Crystal structures of FabI from F. tularensis in complex with novel inhibitors based on the benzimidazole scaffold | | Descriptor: | 1-(4-methoxy-3-methylbenzyl)-1,5,6,7-tetrahydroindeno[5,6-d]imidazole, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Mehboob, S, Boci, T, Brubaker, L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2013-02-01 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and biological evaluation of a novel series of benzimidazole inhibitors of Francisella tularensis enoyl-ACP reductase (FabI).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1O01
 
 | | Human mitochondrial aldehyde dehydrogenase complexed with crotonaldehyde, NAD(H) and Mg2+ | | Descriptor: | (2E)-BUT-2-ENAL, 1,2-ETHANEDIOL, Aldehyde dehydrogenase, ... | | Authors: | Perez-Miller, S.J, Hurley, T.D. | | Deposit date: | 2003-02-20 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Coenzyme isomerization is integral to catalysis in aldehyde dehydrogenase
Biochemistry, 42, 2003
|
|
4IUO
 
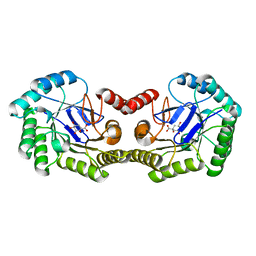 | | 1.8 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) K170M Mutant in Complex with Quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
5GGI
 
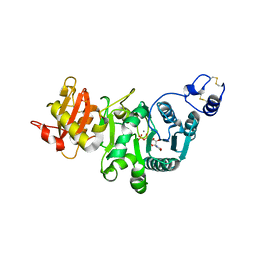 | | Crystal structure of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Mn, UDP and Mannosyl-peptide | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1, ... | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6A36
 
 | | Mandelate oxidase mutant-Y128F with the 3-fluoropyruvic acid FMN adduct | | Descriptor: | 1-deoxy-1-{5-[(1S)-2-fluoro-1-hydroxyethyl]-7,8-dimethyl-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl}-5-O-phosphono-D-ribitol, 1-{5-[(3S)-3-carboxy-4-fluoro-3-hydroxybutanoyl]-7,8-dimethyl-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl}-1-deoxy-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural and chemical trapping of flavin-oxide intermediates reveals substrate-directed reaction multiplicity.
Protein Sci., 29, 2020
|
|
6TEC
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Xylose | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
6TES
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Glucuronic Acid | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
4JMV
 
 | |
4BL5
 
 | | Crystal structure of human GDP-L-fucose synthase with bound NADP and product GDP-L-fucose | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-FUCOSE SYNTHASE, GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, ... | | Authors: | Vollmar, M, Shafqat, N, Rojkova, A, Krojer, T, Bradley, A, Raynor, J.W, Kavanagh, K, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U, Yue, W.W. | | Deposit date: | 2013-05-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Human Gdp-L-Fucose Synthase with Bound Nadp and Product Gdp-L-Fucose
To be Published
|
|
7JH0
 
 | | Crystallographic structure of glyceraldehyde-3-phosphate dehydrogenase from Schistosoma mansoni | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Boreiko, S, Silva, M, Iulek, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure determination and analyses of the GAPDH from the parasite Schistosoma mansoni, the first one from a platyhelminth.
Biochimie, 184, 2021
|
|
4NJO
 
 | |
5DKE
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 3-naphthyl-substituted, methyl, cis-diaryl-ethylene compound 4,4'-[2-(naphthalen-2-yl)prop-1-ene-1,1-diyl]diphenol | | Descriptor: | 4,4'-[2-(naphthalen-2-yl)prop-1-ene-1,1-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DL4
 
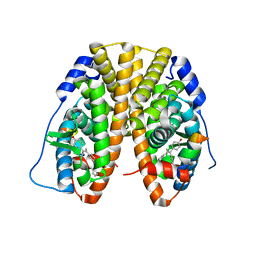 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a phenylamino-substituted, methyl, triaryl-ethylene derivative 4,4'-{2-[3-(phenylamino)phenyl]prop-1-ene-1,1-diyl}diphenol | | Descriptor: | 4,4'-{2-[3-(phenylamino)phenyl]prop-1-ene-1,1-diyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4B49
 
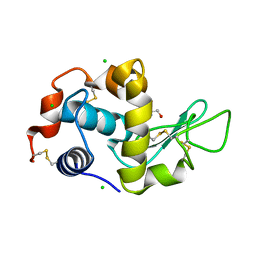 | | 1.15 A Structure of Lysozyme Crystallized without 2-methyl-2,4- pentanediol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, LYSOZYME C | | Authors: | Jakoncic, J, Berger, J, Stauber, M, Axelbaum, A, Asherie, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallization of Lysozyme with (R)-, (S)- and (Rs)-2-Methyl-2,4-Pentanediol
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3VTF
 
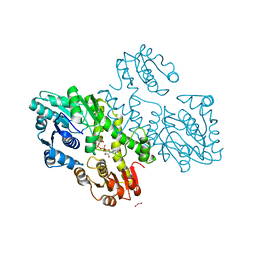 | | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Sakuraba, H, Ohshima, T, Yoneda, K. | | Deposit date: | 2012-05-29 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1E5V
 
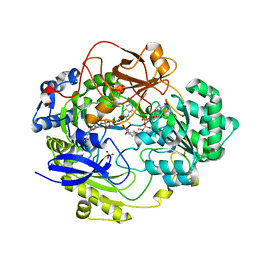 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
2EFZ
 
 | |
6L3R
 
 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 4-bromo-N-((1S,2R)-2-(naphthalen-1-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)benzenesulfonamide | | Descriptor: | 4-bromo-N-((1S,2R)-2-(naphthalen-1-yl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)benzenesulfonamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
6EV2
 
 | | Crystal structure of antibody against schizophyllan in complex with laminarihexaose | | Descriptor: | Heavy chain, Light chain, beta-D-glucopyranose, ... | | Authors: | Sung, K.H, Josewski, J, Duebel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
1VBI
 
 | |
