5NYQ
 
 | |
5NYG
 
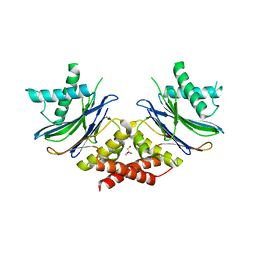 | |
5NYR
 
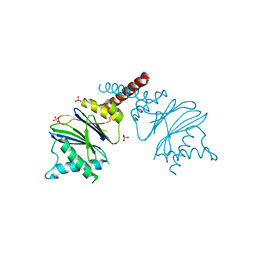 | | Anbu from Hyphomicrobium sp. strain MC1 -SG: R3 | | Descriptor: | Anbu, SULFATE ION | | Authors: | Vielberg, M.-T, Groll, M. | | Deposit date: | 2017-05-11 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | On the Trails of the Proteasome Fold: Structural and Functional Analysis of the Ancestral beta-Subunit Protein Anbu.
J. Mol. Biol., 430, 2018
|
|
4NPR
 
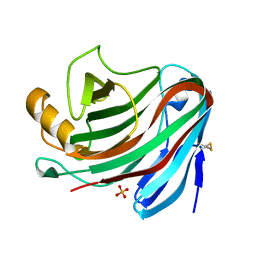 | | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus | | Descriptor: | SULFATE ION, Xyloglucan-specific endo-beta-1,4-glucanase GH12 | | Authors: | Cordeiro, R.L, Santos, C.R, Furtado, G.P, Damasio, A.R.L, Polizeli, M.L.T.M, Ward, R.J, Murakami, M.T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus
To be Published
|
|
4NDI
 
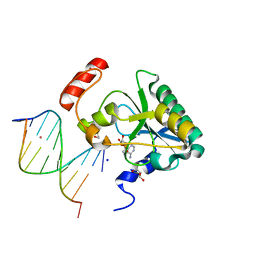 | | Human Aprataxin (Aptx) AOA1 variant K197Q bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDH
 
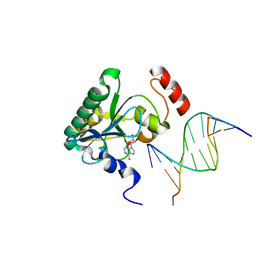 | | Human Aprataxin (Aptx) bound to DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(P*GP*TP*TP*CP*TP*AP*GP*AP*AP*C)-3', ADENOSINE MONOPHOSPHATE, Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDG
 
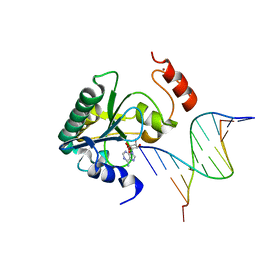 | | Human Aprataxin (Aptx) bound to RNA-DNA and Zn - adenosine vanadate transition state mimic complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4NDF
 
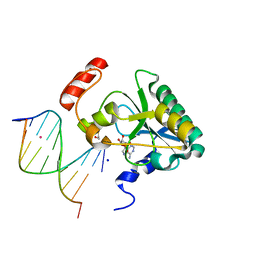 | | Human Aprataxin (Aptx) bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4G1I
 
 | |
4J1B
 
 | |
4J1A
 
 | |
4G08
 
 | |
5OPF
 
 | | Structure of LPMO10B from from Micromonospora aurantiaca | | Descriptor: | COPPER (II) ION, Chitin-binding domain 3 protein | | Authors: | Forsberg, Z, Bissaro, B, Gullesen, J, Dalhus, B, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | Structural determinants of bacterial lytic polysaccharide monooxygenase functionality.
J. Biol. Chem., 293, 2018
|
|
4K8Z
 
 | | KOD Polymerase in binary complex with dsDNA | | Descriptor: | 1,2-ETHANEDIOL, COBALT HEXAMMINE(III), DNA (5'-D(*AP*AP*AP*TP*TP*CP*GP*CP*AP*GP*TP*TP*CP*GP*CP*G)-3'), ... | | Authors: | Bergen, K, Betz, K, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of KOD and 9N DNA Polymerases Complexed with Primer Template Duplex
Chembiochem, 14, 2013
|
|
4LS5
 
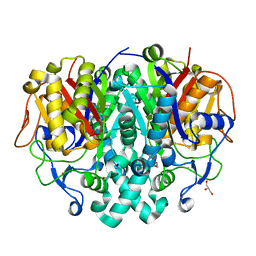 | |
5NPL
 
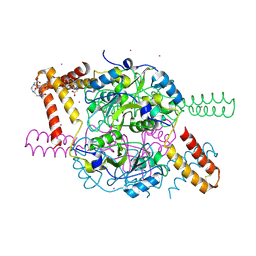 | | Crystal structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria, Yb-derivative at 2.8 A resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, Similar to tr|Q8YYT1|Q8YYT1, YTTERBIUM (III) ION | | Authors: | Hackenberg, C, Hakanpaa, J, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-04-17 | | Release date: | 2018-05-30 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4LS8
 
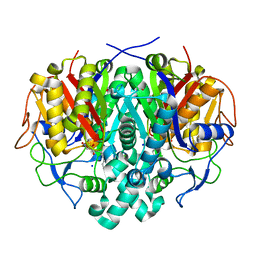 | | Crystal structure of Bacillus subtilis beta-ketoacyl-ACP synthase II (FabF) in a covalent complex with cerulenin | | Descriptor: | (3R,7E,10E)-3-hydroxy-4-oxododeca-7,10-dienamide, 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-07-22 | | Release date: | 2014-04-02 | | Last modified: | 2014-06-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into bacterial resistance to cerulenin.
Febs J., 281, 2014
|
|
4IAU
 
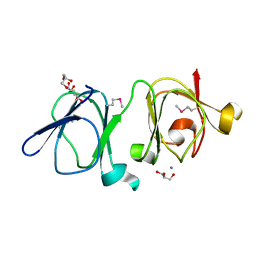 | | Atomic resolution structure of Geodin, a beta-gamma crystallin from Geodia cydonium | | Descriptor: | Beta-gamma-crystallin, CALCIUM ION, GLYCEROL | | Authors: | Vergara, A, Grassi, M, Sica, F, Mazzarella, L, Merlino, A. | | Deposit date: | 2012-12-07 | | Release date: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A novel interdomain interface in crystallins: structural characterization of the [beta][gamma]-crystallin from Geodia cydonium at 0.99 A resolution
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5N7J
 
 | |
4L55
 
 | |
4KZV
 
 | | Structure of the carbohydrate-recognition domain of the C-type lectin mincle bound to trehalose | | Descriptor: | C-type lectin mincle, CALCIUM ION, SODIUM ION, ... | | Authors: | Feinberg, H, Jegouzo, S.A.F, Rowntree, T.J.W, Guan, Y, Brash, M.A, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2013-05-30 | | Release date: | 2013-08-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism for Recognition of an Unusual Mycobacterial Glycolipid by the Macrophage Receptor Mincle.
J.Biol.Chem., 288, 2013
|
|
7KDN
 
 | |
4LS7
 
 | | Crystal structure of Bacillus subtilis beta-ketoacyl-ACP synthase II (FabF) in a non-covalent complex with cerulenin | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, Cerulenin, GLYCEROL, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-07-22 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.674 Å) | | Cite: | Structural insights into bacterial resistance to cerulenin.
Febs J., 281, 2014
|
|
4GZ0
 
 | |
4KZW
 
 | | Structure of the carbohydrate-recognition domain of the C-type lectin mincle | | Descriptor: | C-TYPE LECTIN MINCLE, CALCIUM ION, CITRATE ANION, ... | | Authors: | Feinberg, H, Jegouzo, S.A.F, Rowntree, T.J.W, Guan, Y, Brash, M.A, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2013-05-30 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism for Recognition of an Unusual Mycobacterial Glycolipid by the Macrophage Receptor Mincle.
J.Biol.Chem., 288, 2013
|
|
