7XJW
 
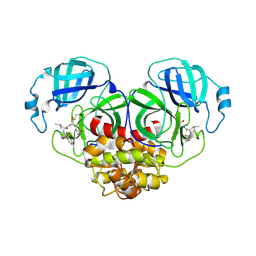 | | Crystal structure of canine coronavirus main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, ORF1a polyprotein | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chiu, Y.F, Chen, Y. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Structural Comparison of SARS-CoV-2 Main Protease and Animal Coronaviral Main Protease Reveals Species-Specific Ligand Binding and Dimerization Mechanism.
Int J Mol Sci, 23, 2022
|
|
7TN1
 
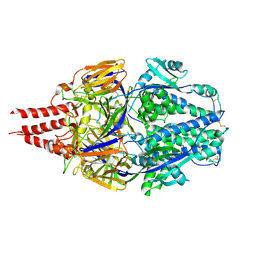 | | Multistate design to stabilize viral class I fusion proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | Huang, J, Banerjee, A, Gonzalez, K, Mousa, J, Strauch, E. | | Deposit date: | 2022-01-20 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Multistate design to stabilize viral class I fusion proteins
To Be Published
|
|
2K0J
 
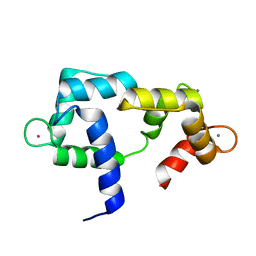 | | Solution structure of CaM complexed to DRP1p | | Descriptor: | CALCIUM ION, LANTHANUM (III) ION, calmodulin | | Authors: | Bertini, I, Luchinat, C, Parigi, G, Yuan, J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-02-04 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Accurate solution structures of proteins from X-ray data and a minimal set of NMR data: calmodulin-peptide complexes as examples.
J.Am.Chem.Soc., 131, 2009
|
|
4JF7
 
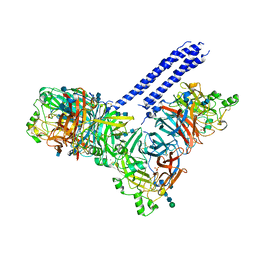 | | Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Welch, B.D, Yuan, P, Bose, S, Kors, C.A, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5018 Å) | | Cite: | Structure of the Parainfluenza Virus 5 (PIV5) Hemagglutinin-Neuraminidase (HN) Ectodomain.
Plos Pathog., 9, 2013
|
|
1G20
 
 | | MGATP-BOUND AND NUCLEOTIDE-FREE STRUCTURES OF A NITROGENASE PROTEIN COMPLEX BETWEEN LEU127DEL-FE PROTEIN AND THE MOFE PROTEIN | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Chiu, H.-J, Peters, J.W, Lanzilotta, W.N, Ryle, M.J, Seefeldt, L.C, Howard, J.B, Rees, D.C. | | Deposit date: | 2000-10-16 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MgATP-Bound and nucleotide-free structures of a nitrogenase protein complex between the Leu 127 Delta-Fe-protein and the MoFe-protein.
Biochemistry, 40, 2001
|
|
4QVJ
 
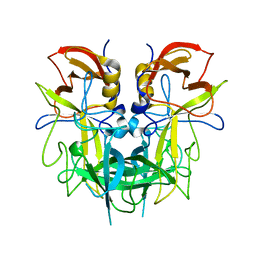 | |
6PY8
 
 | | Crystal structure of the RBPJ-NOTCH1-NRARP ternary complex bound to DNA | | Descriptor: | DNA, Neurogenic locus notch homolog protein 1, Notch-regulated ankyrin repeat-containing protein, ... | | Authors: | Jarrett, S.M, Seegar, T.C.M, Blacklow, S.C. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Extension of the Notch intracellular domain ankyrin repeat stack by NRARP promotes feedback inhibition of Notch signaling.
Sci.Signal., 12, 2019
|
|
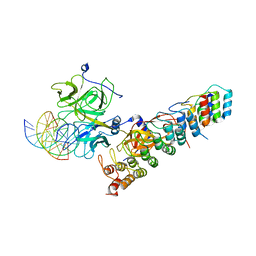
7X1G
 
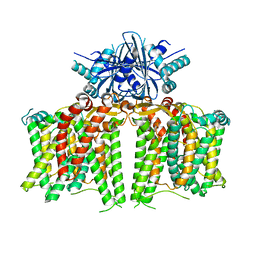 | |
4CYA
 
 | |
7X1J
 
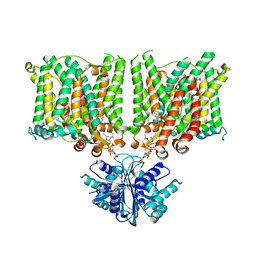 | | Cryo-EM structure of human BTR1 in the outward-facing state in the presence of NH4Cl. | | Descriptor: | Isoform 1 of Solute carrier family 4 member 11, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y, Lu, Y, Zuo, P. | | Deposit date: | 2022-02-24 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into the conformational changes of BTR1/SLC4A11 in complex with PIP 2.
Nat Commun, 14, 2023
|
|
8OPX
 
 | |
7X1H
 
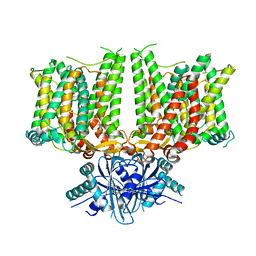 | |
7X1I
 
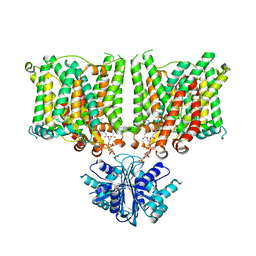 | | Cryo-EM structure of human BTR1 in the outward-facing state. | | Descriptor: | Isoform 1 of Solute carrier family 4 member 11, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y, Lu, Y, Zuo, P. | | Deposit date: | 2022-02-24 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural insights into the conformational changes of BTR1/SLC4A11 in complex with PIP 2.
Nat Commun, 14, 2023
|
|
8DPB
 
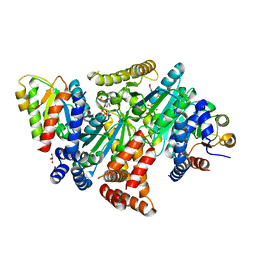 | | MeaB in complex with the cobalamin-binding domain of its target mutase with GMPPCP bound | | Descriptor: | GLYCEROL, MAGNESIUM ION, Methylmalonyl-CoA mutase accessory protein, ... | | Authors: | Vaccaro, F.A, Born, D.A, Drennan, C.L. | | Deposit date: | 2022-07-15 | | Release date: | 2023-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of metallochaperone in complex with the cobalamin-binding domain of its target mutase provides insight into cofactor delivery.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8OQO
 
 | |
3R24
 
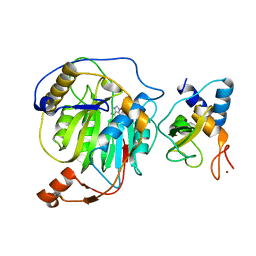 | | Crystal structure of nsp10/nsp16 complex of SARS coronavirus | | Descriptor: | 2'-O-methyl transferase, Non-structural protein 10 and Non-structural protein 11, S-ADENOSYLMETHIONINE, ... | | Authors: | Liu, X, Guo, D, Su, C, Chen, Y. | | Deposit date: | 2011-03-13 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex.
Plos Pathog., 7, 2011
|
|
7XFJ
 
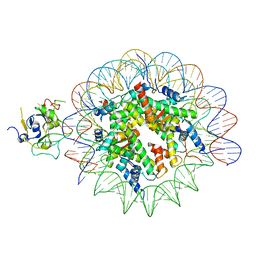 | | Structure of nucleosome-AAG complex (T-50I, post-catalytic state) | | Descriptor: | DNA (152-MER), DNA-3-methyladenine glycosylase, Histone H2A type 1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
8OQL
 
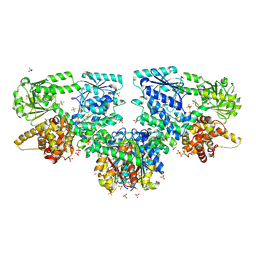 | |
8OPU
 
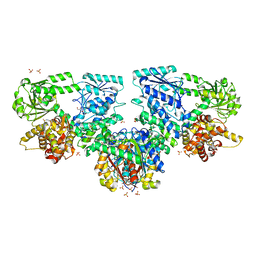 | |
8OPW
 
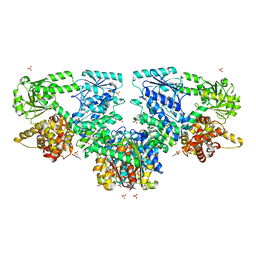 | |
8OPY
 
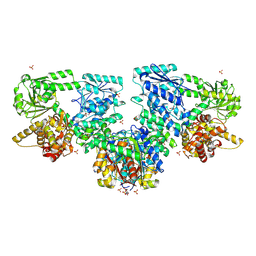 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-B-DNQ | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, 6,7-DINITROQUINOXALINE-2,3-DIONE, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystallographic fragment binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate channeling path between them
Biorxiv, 2024
|
|
8OQN
 
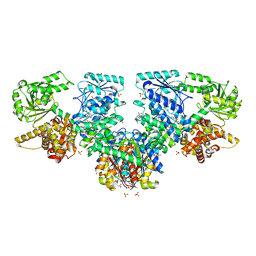 | |
4R9L
 
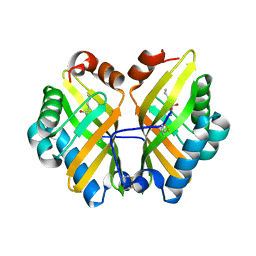 | | Structure of a thermostable elevenfold mutant of limonene epoxide hydrolase from Rhodococcus erythropolis, containing two stabilizing disulfide bonds | | Descriptor: | (2R)-2-hydroxyhexanamide, Limonene-1,2-epoxide hydrolase | | Authors: | Floor, R.J, Wijma, H.J, Jekel, P.A, Terwisscha van Scheltinga, A.C, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Proteins, 83, 2015
|
|
8OPV
 
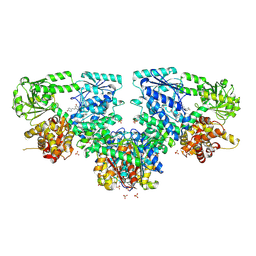 | |
8OQM
 
 | |
