1EL4
 
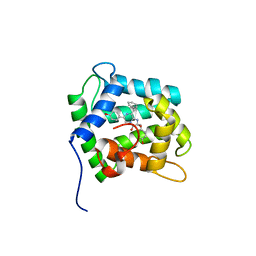 | | STRUCTURE OF THE CALCIUM-REGULATED PHOTOPROTEIN OBELIN DETERMINED BY SULFUR SAS | | Descriptor: | C2-HYDROXY-COELENTERAZINE, CHLORIDE ION, OBELIN | | Authors: | Liu, Z.J, Vysotski, E.S, Rose, J, Lee, J, Wang, B.C. | | Deposit date: | 2000-03-13 | | Release date: | 2001-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of the Ca2+-regulated photoprotein obelin at 1.7 A resolution determined directly from its sulfur substructure.
Protein Sci., 9, 2000
|
|
6UVR
 
 | |
6V4J
 
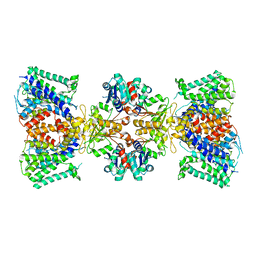 | | Structure of TrkH-TrkA in complex with ATP | | Descriptor: | Potassium uptake protein TrkA, Trk system potassium uptake protein TrkH | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
6UVT
 
 | |
6YAI
 
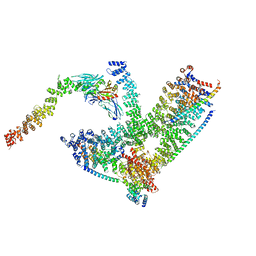 | | Clathrin with bound beta2 appendage of AP2 | | Descriptor: | AP-2 complex subunit beta, Clathrin heavy chain, Clathrin light chain | | Authors: | Kovtun, O, Kane Dickson, V, Kelly, B.T, Owen, D, Briggs, J.A.G. | | Deposit date: | 2020-03-12 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Architecture of the AP2/clathrin coat on the membranes of clathrin-coated vesicles.
Sci Adv, 6, 2020
|
|
8SCY
 
 | | Lysozyme crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | Lysozyme C | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6PCI
 
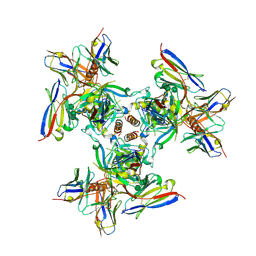 | | EBOV GPdMuc (Makona) in complex with rEBOV-520 and rEBOV-548 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Virion spike glycoprotein, Virion spike glycoprotein,Virion spike glycoprotein,Ebola Virus (Makona) GP2, ... | | Authors: | Ward, A.B, Murin, C.D, Alkutkar, T. | | Deposit date: | 2019-06-17 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization.
Immunity, 52, 2020
|
|
6MW9
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-3 antibody | | Descriptor: | E1, E2, EEEV-3 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6W6P
 
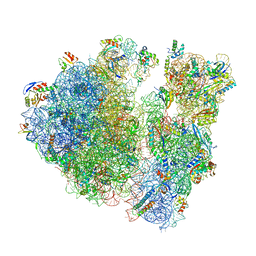 | |
8C8R
 
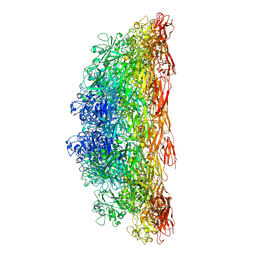 | |
6MUI
 
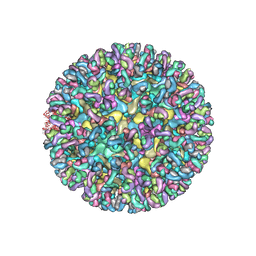 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-42 antibody | | Descriptor: | E1, E2, EEEV-42 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6T7D
 
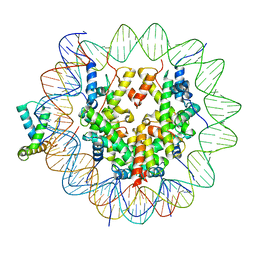 | | Structure of human Sox11 transcription factor in complex with a nucleosome | | Descriptor: | DNA (151-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Dodonova, S.O, Zhu, F, Dienemann, C, Taipale, J, Cramer, P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function.
Nature, 580, 2020
|
|
6T79
 
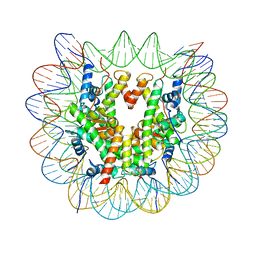 | | Structure of a human nucleosome at 3.2 A resolution | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Dodonova, S.O, Zhu, F, Dienemann, C, Taipale, J, Cramer, P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function.
Nature, 580, 2020
|
|
8SIL
 
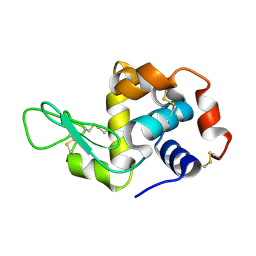 | | Lysozyme crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | Lysozyme C | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L, Botha, S, Poitevin, F, Sierra, R.G. | | Deposit date: | 2023-04-16 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5ON2
 
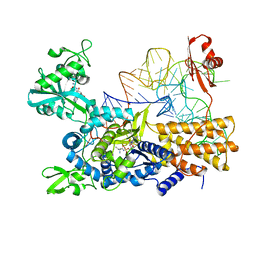 | | Quaternary complex of mutant T252A of E. coli leucyl-tRNA synthetase with tRNA(leu), leucyl-adenylate analogue, and post-transfer editing analogue of norvaline in the aminoacylation conformation | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | Authors: | Palencia, A, Cusack, S. | | Deposit date: | 2017-08-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Kinetic Origin of Substrate Specificity in Post-Transfer Editing by Leucyl-tRNA Synthetase.
J. Mol. Biol., 430, 2018
|
|
5OX3
 
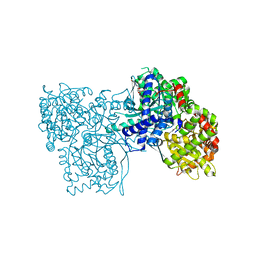 | | Glycogen Phosphorylase in complex with SzB102v | | Descriptor: | (1S)-1,5-anhydro-1-[3-(4-hydroxyphenyl)-1H-1,2,4-triazol-5-yl]-D-glucitol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Kantsadi, A.L, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the beta-pocket of the active site of human liver glycogen phosphorylase with 3-(C-beta-d-glucopyranosyl)-5-(4-substituted-phenyl)-1, 2, 4-triazole inhibitors.
Bioorg. Chem., 77, 2018
|
|
6MW3
 
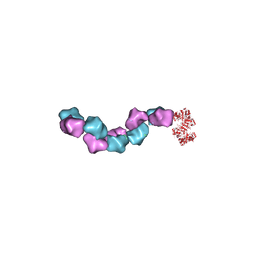 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited filament composed of NrdE alpha subunit and NrdF beta subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase, Ribonucleoside-diphosphate reductase NrdF beta subunit | | Authors: | Thomas, W.C, Bacik, J.P, Kaelber, J.T, Ando, N. | | Deposit date: | 2018-10-29 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
5OMW
 
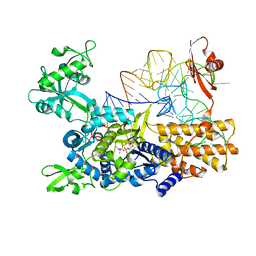 | |
5ONH
 
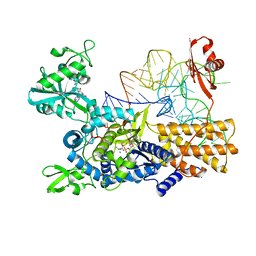 | | Quaternary complex of wild type E. coli leucyl-tRNA synthetase with tRNA(leu), leucyl-adenylate analogue, and post-transfer editing analogue of norvaline in the aminoacylation conformation | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, L-leucyl-tRNA(Leu), ... | | Authors: | Palencia, A, Cusack, S. | | Deposit date: | 2017-08-03 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Kinetic Origin of Substrate Specificity in Post-Transfer Editing by Leucyl-tRNA Synthetase.
J. Mol. Biol., 430, 2018
|
|
5OWY
 
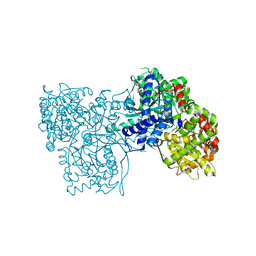 | | Glycogen Phosphorylase in complex with KS252 | | Descriptor: | 4-[5-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-4~{H}-1,2,4-triazol-3-yl]benzoic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stravodimos, G, Kantsadi, A, Chatzileontiadou, D, Leonidas, D. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the beta-pocket of the active site of human liver glycogen phosphorylase with 3-(C-beta-d-glucopyranosyl)-5-(4-substituted-phenyl)-1, 2, 4-triazole inhibitors.
Bioorg. Chem., 77, 2018
|
|
5OX1
 
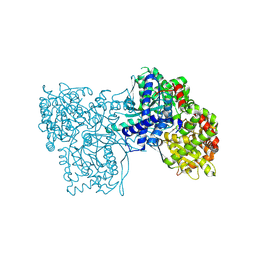 | | Glycogen Phosphorylase in complex with JLH270 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-[5-(4-methoxyphenyl)-1~{H}-1,2,4-triazol-3-yl]oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Kantsadi, A.L, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the beta-pocket of the active site of human liver glycogen phosphorylase with 3-(C-beta-d-glucopyranosyl)-5-(4-substituted-phenyl)-1, 2, 4-triazole inhibitors.
Bioorg. Chem., 77, 2018
|
|
6WGG
 
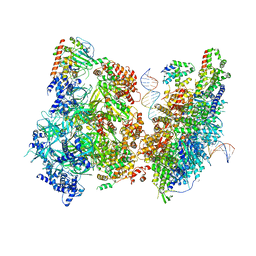 | | Atomic model of pre-insertion mutant OCCM-DNA complex(ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (41-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X9O
 
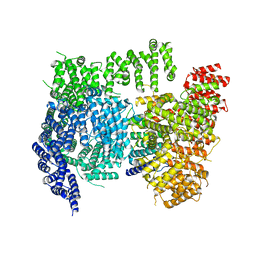 | | High resolution cryoEM structure of huntingtin in complex with HAP40 | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin | | Authors: | Harding, R.J, Deme, J.C, Lea, S.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Huntingtin structure is orchestrated by HAP40 and shows a polyglutamine expansion-specific interaction with exon 1.
Commun Biol, 4, 2021
|
|
8FD5
 
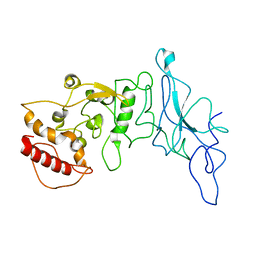 | | Nucleocapsid monomer structure from SARS-CoV-2 | | Descriptor: | Nucleoprotein | | Authors: | Casasanta, M, Jonaid, G.M, Kaylor, L, Luqiu, W, DiCecco, L, Solares, M, Berry, S, Kelly, D.F. | | Deposit date: | 2022-12-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (4.57 Å) | | Cite: | Structural Insights of the SARS-CoV-2 Nucleocapsid Protein: Implications for the Inner-workings of Rapid Antigen Tests.
Microsc Microanal, 29, 2023
|
|
8FG2
 
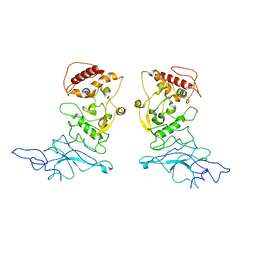 | | SARS-CoV-2 Nucleocapsid dimer structure determined from COVID-19 patients | | Descriptor: | Nucleoprotein | | Authors: | Casasanta, M, Jonaid, G.M, Kaylor, L, Luqiu, W, DiCecco, L, Solares, M, Berry, S, Kelly, D.F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural Insights of the SARS-CoV-2 Nucleocapsid Protein: Implications for the Inner-workings of Rapid Antigen Tests.
Microsc Microanal, 29, 2023
|
|
