5KVQ
 
 | |
7NE6
 
 | | Human TET2 in complex with unfavourable DNA substrate. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*CP*AP*GP*GP*(5CM)P*GP*CP*CP*TP*G)-3'), ... | | Authors: | Rafalski, D, Bochtler, M. | | Deposit date: | 2021-02-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pronounced sequence specificity of the TET enzyme catalytic domain guides its cellular function.
Sci Adv, 8, 2022
|
|
6EP3
 
 | | Lar controls the expression of the Listeria monocytogenes agr system and mediates virulence. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lmo0651 protein, ... | | Authors: | Pinheiro, J, Lisboa, J, Carvalho, F, Carreaux, A, Santos, N, Cabral, J, Sousa, S, Cabanes, D. | | Deposit date: | 2017-10-10 | | Release date: | 2018-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MouR controls the expression of the Listeria monocytogenes Agr system and mediates virulence.
Nucleic Acids Res., 46, 2018
|
|
6JTD
 
 | | Crystal structure of TcCGT1 in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, C-glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhao, P, Yun, C.H. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular and Structural Characterization of a Promiscuous C-Glycosyltransferase from Trollius chinensis.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5O1N
 
 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with N-[(2S)-2-Pyrrolidinylmethyl]-trifluoromethanesulfonamide bound | | Descriptor: | 1,1,1-tris(fluoranyl)-~{N}-[[(2~{S})-pyrrolidin-2-yl]methyl]methanesulfonamide, 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Strain-Damerell, C, Goubin, S, Sethi, R, Velupillai, S, Talon, R, Collins, P, Krojer, T, McLaughlin, M, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with N-[(2S)-2-Pyrrolidinylmethyl]-trifluoromethanesulfonamide bound
To Be Published
|
|
7N9I
 
 | | 1.4A Structure of Drosophila melanogaster Frataxin | | Descriptor: | Frataxin homolog, mitochondrial, SULFATE ION | | Authors: | Brunzelle, J.S, Stemmler, T.L. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Drosophila melanogaster frataxin: protein crystal and predicted solution structure with identification of the iron-binding regions.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
9KEB
 
 | | Crystal structure of the PIN1 and fragment 24 complex. | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-11-04 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
5I9W
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with ANP | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.359 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
6FXQ
 
 | | Structure of coproheme decarboxylase from Listeria monocytogenes during turnover | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | Authors: | Hofbauer, S, Pfanzagl, V, Mlynek, G, Puehringer, D. | | Deposit date: | 2018-03-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Redox Cofactor Rotates during Its Stepwise Decarboxylation: Molecular Mechanism of Conversion of Coproheme to Hemeb.
Acs Catalysis, 9, 2019
|
|
6BXG
 
 | | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide. | | Descriptor: | CHLORIDE ION, IODIDE ION, LEU-ILE-ALA, ... | | Authors: | Minasov, G, Shuvalova, L, Filippova, E.V, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-18 | | Release date: | 2018-01-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide.
To Be Published
|
|
5ILG
 
 | | Crystal structure of photoreceptor dehydrogenase from Drosophila melanogaster | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hofmann, L, Tsybovsky, Y, Banerjee, S. | | Deposit date: | 2016-03-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family.
Biochemistry, 55, 2016
|
|
1QMZ
 
 | | PHOSPHORYLATED CDK2-CYCLYIN A-SUBSTRATE PEPTIDE COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, G2/MITOTIC-SPECIFIC CYCLIN A, ... | | Authors: | Brown, N.R, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 1999-10-11 | | Release date: | 1999-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for Specificity of Substrate and Recruitment Peptides for Cyclin-Dependent Kinases
Nat.Cell Biol., 1, 1999
|
|
6MUK
 
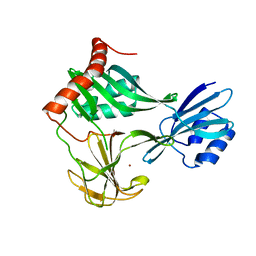 | | 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae. | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-23 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae.
To Be Published
|
|
3M3H
 
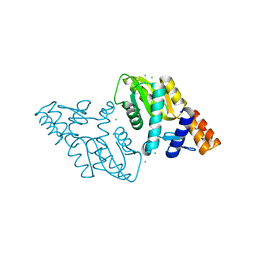 | | 1.75 Angstrom resolution crystal structure of an orotate phosphoribosyltransferase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | CHLORIDE ION, Orotate phosphoribosyltransferase | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Winsor, J, Dubrovska, I, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-09 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom resolution crystal structure of an orotate phosphoribosyltransferase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
3MA9
 
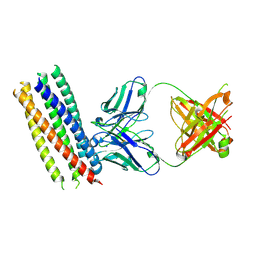 | | Crystal structure of gp41 derived protein complexed with fab 8066 | | Descriptor: | Fab8066 FAB ANTIBODY FRAGMENT, Heavy Chain, Light Chain, ... | | Authors: | Li, M, Gustchina, E, Louis, J, Gustchina, A, Wlodawer, A, Clore, M. | | Deposit date: | 2010-03-23 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41.
Plos Pathog., 6, 2010
|
|
5H5Q
 
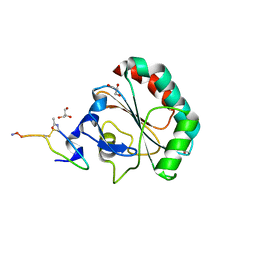 | | Crystal structure of human GPX4 in complex with GXpep-1 | | Descriptor: | GLYCEROL, GXpep-1, Phospholipid hydroperoxide glutathione peroxidase, ... | | Authors: | Sogabe, S, Kadotani, A, Lane, W, Snell, G. | | Deposit date: | 2016-11-09 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery of GPX4 inhibitory peptides from random peptide T7 phage display and subsequent structural analysis
Biochem. Biophys. Res. Commun., 482, 2017
|
|
6DJE
 
 | | Crystal structure of Tdp1 catalytic domain in complex with Sigma Aldrich compound CDS010292 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-8-(propan-2-yl)quinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DOU
 
 | |
5A1T
 
 | | Trichomonas vaginalis lactate dehydrogenase in complex with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-LACTATE DEHYDROGENASE, OXAMIC ACID | | Authors: | Steindel, P.A, Chen, E.H, Theobald, D.L. | | Deposit date: | 2015-05-05 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Gradual Neofunctionalization in the Convergent Evolution of Trichomonad Lactate and Malate Dehydrogenases.
Protein Sci., 25, 2016
|
|
6D8E
 
 | |
2WCZ
 
 | | 1.6A resolution structure of Archaeoglobus fulgidus Hjc, a Holliday junction resolvase from an archaeal hyperthermophile | | Descriptor: | CHLORIDE ION, HOLLIDAY JUNCTION RESOLVASE | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1.6 A Resolution Structure of Archaeoglobus Fulgidus Hjc, a Holliday Junction Resolvase from an Archaeal Hyperthermophile
To be Published
|
|
5IO3
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - I422 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
6FQO
 
 | | Crystal structure of CREBBP bromodomain complexd with DT29 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, ~{N}-[3-(2,5-dimethyl-3-oxidanylidene-1,2-oxazol-4-yl)-5-(5-ethanoyl-2-ethoxy-phenyl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6G4M
 
 | | Torpedo californica acetylcholinesterase bound to uncharged hybrid reactivator 1 | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-[4-(1,2,3,4-tetrahydroacridin-9-ylamino)butyl]pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
3MUK
 
 | |
