7E1N
 
 | | Crystal structure of PhlH in complex with 2,4-diacetylphloroglucinol | | Descriptor: | 2,4-bis[(1R)-1-oxidanylethyl]benzene-1,3,5-triol, DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
5YDV
 
 | | Regulatory domain of HypT from Salmonella typhimurium complexed with HOCl (HOCl-bound form) | | Descriptor: | Cell density-dependent motility repressor, SULFATE ION, hypochlorous acid | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YEZ
 
 | | Regulatory domain of HypT M206Q mutant from Salmonella typhimurium | | Descriptor: | Cell density-dependent motility repressor | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6N2G
 
 | | Crystal structure of Caenorhabditis elegans NAP1 | | Descriptor: | Nucleosome Assembly Protein | | Authors: | Bhattacharyya, S, DArcy, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Characterization of Caenorhabditis elegans Nucleosome Assembly Protein 1 Uncovers the Role of Acidic Tails in Histone Binding.
Biochemistry, 58, 2019
|
|
5YER
 
 | | Regulatory domain of HypT from Salmonella typhimurium (Bromide ion-bound) | | Descriptor: | BROMIDE ION, Cell density-dependent motility repressor, SULFATE ION | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-19 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YDO
 
 | | Regulatory domain of HypT from Salmonella typhimurium (apo-form) | | Descriptor: | Cell density-dependent motility repressor, SULFATE ION | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-13 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4P96
 
 | |
6PV1
 
 | |
6NT5
 
 | | Cryo-EM structure of full-length human STING in the apo state | | Descriptor: | Stimulator of interferon protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6CBI
 
 | | PCNA in complex with inhibitor | | Descriptor: | GLY-ARG-LYS-ARG-ARG-GLN-DAB-SER-MET-THR-GLU-PHE-TYR-HIS, Proliferating cell nuclear antigen, SULFATE ION | | Authors: | Bruning, J.B, Wegener, K.L. | | Deposit date: | 2018-02-03 | | Release date: | 2018-07-04 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rational Design of a 310-Helical PIP-Box Mimetic Targeting PCNA, the Human Sliding Clamp.
Chemistry, 24, 2018
|
|
6PV3
 
 | |
6G16
 
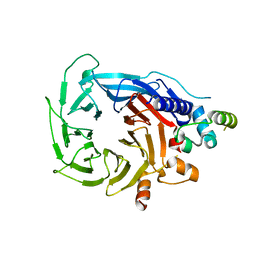 | | Structure of the human RBBP4:MTA1(464-546) complex showing loop exchange | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
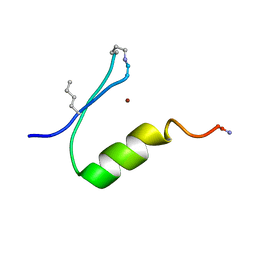
6PV2
 
 | |
6PV0
 
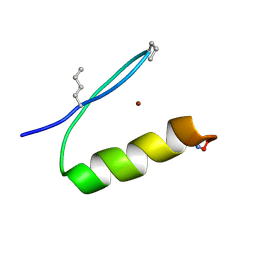 | |
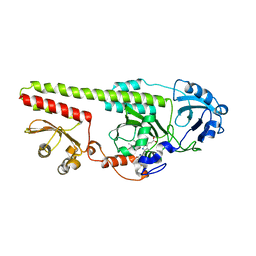
5OY5
 
 | |
7LPS
 
 | | Crystal structure of DDB1-CRBN-ALV1 complex bound to Helios (IKZF2 ZF2) | | Descriptor: | 3-[3-[[1-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2,5-bis(oxidanylidene)pyrrol-3-yl]amino]phenyl]-~{N}-(3-chloranyl-4-methyl-phenyl)propanamide, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, Fischer, E.S. | | Deposit date: | 2021-02-12 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Acute pharmacological degradation of Helios destabilizes regulatory T cells.
Nat.Chem.Biol., 17, 2021
|
|
2H3C
 
 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
4AC6
 
 | | Corynebacterium glutamicum AcnR AU derivative structure | | Descriptor: | GOLD ION, HTH-TYPE TRANSCRIPTIONAL REPRESSOR ACNR | | Authors: | Garcia-Nafria, J, Baumgart, M, Turkenburg, J.P, Wilkinson, A.J, Bott, M, Wilson, K.S. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal and Solution Studies Reveal that the Transcriptional Regulator Acnr of Corynebacterium Glutamicum is Regulated by Citrate:Mg2+ Binding to a Non-Canonical Pocket.
J.Biol.Chem., 288, 2013
|
|
2H3A
 
 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
7WZE
 
 | |
4AF5
 
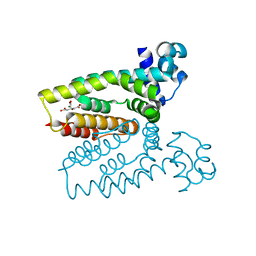 | | Structure of the C. glutamicum AcnR Crystal Form I | | Descriptor: | CITRIC ACID, HTH-TYPE TRANSCRIPTIONAL REPRESSOR ACNR COMPND 2, MAGNESIUM ION | | Authors: | Garcia-Nafria, J, Baumgart, M, Turkenburg, J.P, Wilkinson, A.J, Bott, M, Wilson, K.S. | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal and Solution Studies Reveal that the Transcriptional Regulator Acnr of Corynebacterium Glutamicum is Regulated by Citrate:Mg2+ Binding to a Non-Canonical Pocket.
J.Biol.Chem., 288, 2013
|
|
4ACI
 
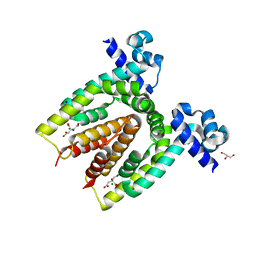 | | Structure of the C. glutamicum AcnR Crystal Form II | | Descriptor: | CITRIC ACID, GLYCEROL, HTH-TYPE TRANSCRIPTIONAL REPRESSOR ACNR, ... | | Authors: | Garcia-Nafria, J, Baumgart, M, Turkenburg, J.P, Wilkinson, A.J, Bott, M, Wilson, K.S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal and Solution Studies Reveal that the Transcriptional Regulator Acnr of Corynebacterium Glutamicum is Regulated by Citrate:Mg2+ Binding to a Non-Canonical Pocket.
J.Biol.Chem., 288, 2013
|
|
4XGC
 
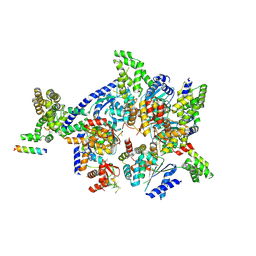 | | Crystal structure of the eukaryotic origin recognition complex | | Descriptor: | CHLORIDE ION, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Bleichert, F, Botchan, M.R, Berger, J.M. | | Deposit date: | 2014-12-30 | | Release date: | 2015-04-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the eukaryotic origin recognition complex.
Nature, 519, 2015
|
|
7PPJ
 
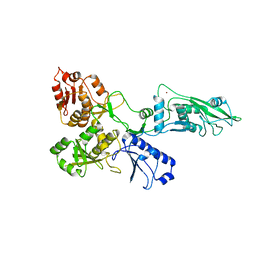 | | human SLFN5 | | Descriptor: | Schlafen family member 5, ZINC ION | | Authors: | Lammens, K, Metzner, F.J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural and biochemical characterization of human Schlafen 5.
Nucleic Acids Res., 50, 2022
|
|
4UP1
 
 | |
