5YAP
 
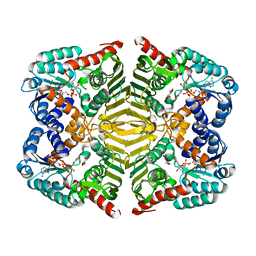 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with L-glucono-1,5-lactone | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
6RMF
 
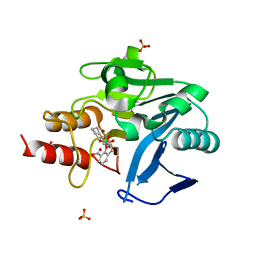 | | Crystal structure of NDM-1 with VNRX-5133 | | Descriptor: | (10aR)-2-(((1r,4R)-4-((2-aminoethyl)amino)cyclohexyl)methyl)-6-carboxy-4-hydroxy-4,10a-dihydro-10H-benzo[5,6][1,2]oxaborinino[2,3-b][1,4,2]oxazaborol-4-uide, (4~{R})-4-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, Metallo-beta-lactamase type 2, ... | | Authors: | Hinchliffe, P, Spencer, J, Brem, J, Schofield, C.J. | | Deposit date: | 2019-05-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Bicyclic Boronate VNRX-5133 Inhibits Metallo- and Serine-beta-Lactamases.
J.Med.Chem., 62, 2019
|
|
6ZAT
 
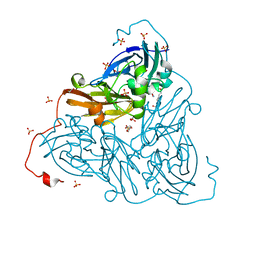 | | Nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at 1.0 A resolution (unrestrained full matrix refinement by SHELX) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
9EKN
 
 | | Thomasclavelia ramosa Immunoglobulin A Protease Middle (Protease) Domain with C-terminal Domain #1 (Metal Chelated and Zinc Supplemented) | | Descriptor: | 1,2-ETHANEDIOL, IgA protease, ZINC ION | | Authors: | Tran, N, Frenette, A, Holyoak, T. | | Deposit date: | 2024-12-02 | | Release date: | 2025-09-03 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Thomasclavelia ramosa immunoglobulin A protease reveals a modular and minimizable architecture distinct from other immunoglobulin A proteases.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
5Y9A
 
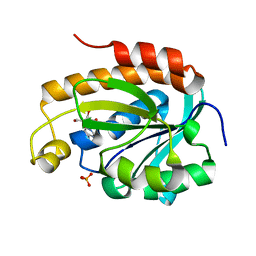 | | Crystal structure of the complex of peptidyl tRNA hydrolase with a phosphate ion at the substrate binding site and cytarabine at a new ligand binding site at 1.1 A resolution | | Descriptor: | CYTARABINE, PHOSPHATE ION, Peptidyl-tRNA hydrolase | | Authors: | Kaushik, S, Iqbal, N, Singh, N, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-23 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Search of multiple hot spots on the surface of peptidyl-tRNA hydrolase: structural, binding and antibacterial studies.
Biochem. J., 475, 2018
|
|
8K17
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to collagen alpha-1(I) chain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, FE (III) ION, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-07-10 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
9JLO
 
 | | Crystal structure L-lactate dehydrogenase from Lactobacillus reuteri in its apoform | | Descriptor: | 1,2-ETHANEDIOL, L-lactate dehydrogenase | | Authors: | Matyuta, I.O, Minyaev, M.E, Shirokova, A.A, Pometun, A.A, Tishkov, V.I, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-09-19 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Novel extremely stable and active lactate dehydrogenase from l reuteri: structure-function studies and promising usage as an agent against pathogenic biofilms
To Be Published
|
|
4Y8K
 
 | | Yeast 20S proteasome in complex with H-APLL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, H-APLL-ep, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
6GHM
 
 | | Structure of PP1 alpha phosphatase bound to ASPP2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Apoptosis-stimulating of p53 protein 2, ... | | Authors: | Mouilleron, S, Bertran, T.M, Tapon, N, Zhou, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | ASPP proteins discriminate between PP1 catalytic subunits through their SH3 domain and the PP1 C-tail.
Nat Commun, 10, 2019
|
|
9RQB
 
 | | Fragment screening of FosAKP, cryo structure in complex with fragment F2X-entry F10 | | Descriptor: | (1~{S})-1-[4-(cyclopropylmethoxy)-3-fluoranyl-phenyl]ethanamine, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2025-06-25 | | Release date: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Room temperature X-ray fragment screening with serial crystallography
To Be Published
|
|
8V8E
 
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease catalytic domain (residues 1-199-6H) in complex with ensitrelvir (ESV) | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, peptide | | Authors: | Kovalevsky, A, Coates, L. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Visualizing the Active Site Oxyanion Loop Transition Upon Ensitrelvir Binding and Transient Dimerization of SARS-CoV-2 Main Protease.
J.Mol.Biol., 436, 2024
|
|
6GV2
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate aldolase Y103F,Y130F,A198F variant in complex with L-2-keto, 3-deoxy-galactonate | | Descriptor: | 1,2-ETHANEDIOL, 2-dehydro-3-deoxy-phosphogluconate/2-dehydro-3-deoxy-6-phosphogalactonate aldolase, 3-DEOXY-D-ARABINO-HEXONIC ACID, ... | | Authors: | Crennell, S.J, Danson, M.J, Royer, S. | | Deposit date: | 2018-06-20 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Sulfolobus solfataricus 2-keto-3-deoxygluconate aldolase Y132V,T157C variant
To Be Published
|
|
8V8G
 
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease catalytic domain (residues 1-196) in complex with ensitrelvir (ESV) | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Kovalevsky, A, Coates, L. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Visualizing the Active Site Oxyanion Loop Transition Upon Ensitrelvir Binding and Transient Dimerization of SARS-CoV-2 Main Protease.
J.Mol.Biol., 436, 2024
|
|
8RZG
 
 | | ZgGH129 from Zobellia galactanivorans soaked with the product of the reaction ADG (3,6-anhydro-D-galactose). | | Descriptor: | (1~{R},4~{S},5~{R},8~{S})-2,6-dioxabicyclo[3.2.1]octane-4,8-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
5I3T
 
 | | Native Structure of the Linalool Dehydratase-Isomerase from Castellaniella defragrans | | Descriptor: | 1,3-BUTANEDIOL, CHLORIDE ION, Linalool dehydratase/isomerase, ... | | Authors: | LaMattina, J.W, Carlock, M, Koch, D.J, Lanzilotta, W.N. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Native Structure of the Linalool Dehydratase-Isomerase from Castellaniella defragrans
To Be Published
|
|
8BFF
 
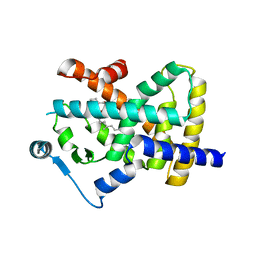 | | Human PPARgamma in complex with MINCH bound to the AF-2 sub-pocket | | Descriptor: | (1~{S},2~{R})-2-[(4~{R})-4-methylheptoxy]carbonylcyclohexane-1-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Useini, A, Straeter, N. | | Deposit date: | 2022-10-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the activation of PPAR gamma by the plasticizer metabolites MEHP and MINCH.
Environ Int, 173, 2023
|
|
6DE2
 
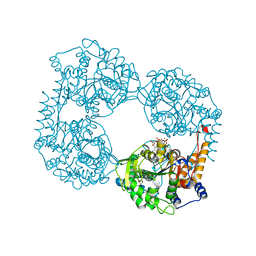 | | Crystal structure of the double mutant (D52N/L375F) of the full-length NT5C2 in the active state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, ... | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
5YSB
 
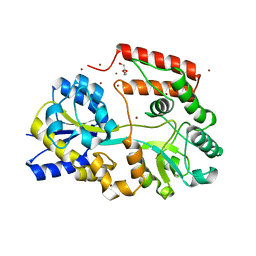 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in ligand-free form | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lin1841 protein, ZINC ION | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5MHA
 
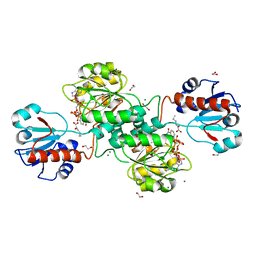 | | D-2-hydroxyacid dehydrogenases (D2-HDH) from Haloferax mediterranei in complex with a mixture of 2-ketohexanoic acid and 2-hydroxyhexanoic acid, and NADPH (1.57 A resolution) | | Descriptor: | (2R)-2-hydroxyhexanoic acid, 1,2-ETHANEDIOL, 2-Ketohexanoic acid, ... | | Authors: | Bisson, C, Baker, P.J, Domenech Perez, J, Pramanpol, N, Harding, S.E, Rice, D.W, Ferrer, J. | | Deposit date: | 2016-11-23 | | Release date: | 2018-05-09 | | Last modified: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Potassium binding by carbonyl clusters, halophilic adaptation and catalysis of Haloferax mediterranei D-2-hydroxyacid dehydrogenase.
Commun Biol, 8, 2025
|
|
8X81
 
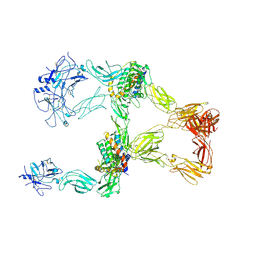 | | Structure of leptin-LepR trimer with a large gap | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Leptin, ... | | Authors: | Xie, Y.F, Gao, G.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-08-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural plasticity of human leptin binding to its receptor LepR
Hlife, 1, 2023
|
|
6ZTW
 
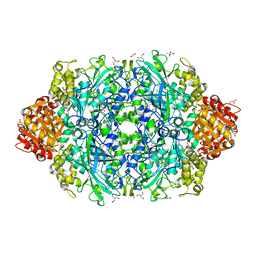 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
5OM6
 
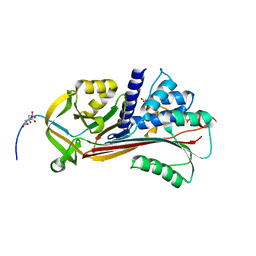 | |
5EUM
 
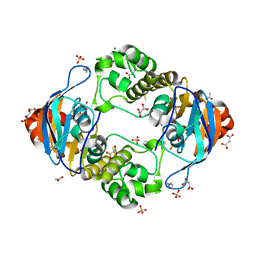 | | 1.8 Angstrom Crystal Structure of ATP-binding Component of Fused Lipid Transporter Subunits of ABC superfamily from Haemophilus influenzae. | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipid A export ATP-binding/permease protein MsbA, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of ATP-binding Component of Fused Lipid Transporter Subunits of ABC superfamily from Haemophilus influenzae.
To Be Published
|
|
6FP9
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_E11 | | Descriptor: | 1,2-ETHANEDIOL, DARPin 1238_E11, SULFATE ION | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
5O1P
 
 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase. | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Williams, E, Velupillai, S, Kupinska, K, Strain-Damerell, C, Goubin, S, Talon, R, Collins, P, Krojer, T, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase.
To Be Published
|
|
