5MF0
 
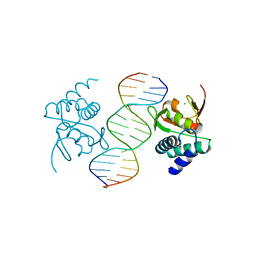 | | Crystal structure of Smad4-MH1 bound to the GGCCG site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*GP*CP*CP*CP*GP*T)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5OAY
 
 | |
5OKC
 
 | |
5MEZ
 
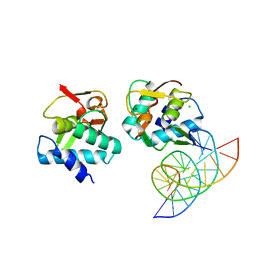 | | Crystal structure of Smad4-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*GP*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5SY5
 
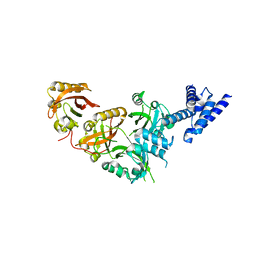 | | Crystal Structure of the Heterodimeric NPAS1-ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Neuronal PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Potluri, N, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors.
Elife, 5, 2016
|
|
7P6F
 
 | | 1.93 A resolution X-ray crystal structure of the transcriptional regulator SrnR from Streptomyces griseus | | Descriptor: | ACETATE ION, SODIUM ION, Transcriptional regulator SrnR | | Authors: | Mazzei, L, Ciurli, S. | | Deposit date: | 2021-07-16 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure, dynamics, and function of SrnR, a transcription factor for nickel-dependent gene expression.
Metallomics, 13, 2021
|
|
5FV7
 
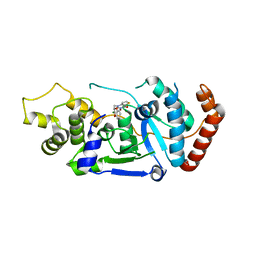 | | Human Fen1 in complex with an N-hydroxyurea compound | | Descriptor: | 1-[(2S)-2,3-dihydro-1,4-benzodioxin-2-ylmethyl]-3-hydroxythieno[3,2-d]pyrimidine-2,4(1H,3H)-dione, FLAP ENDONUCLEASE 1, MAGNESIUM ION | | Authors: | Exell, J.C, Thompson, M.J, Finger, L.D, Shaw, S.K, Abbott, W.M, McWhirter, C, Debreczeni, J.E, Jones, C.D, Nissink, J.W.M, Ward, T.A, Sioberg, C.W.L, Molina, D.M, Durant, S.T, Grasby, J.A. | | Deposit date: | 2016-02-03 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Cellular Active N-Hydroxyurea Fen1 Inhibitors Block Substrate Entry to the Active Site
Nat.Chem.Biol., 12, 2016
|
|
6QKQ
 
 | |
6QKP
 
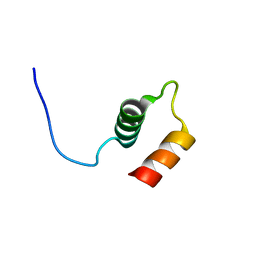 | |
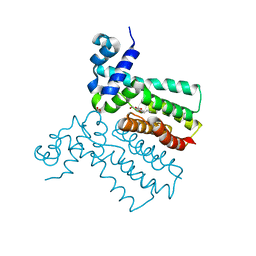
6SRN
 
 | |
8IQG
 
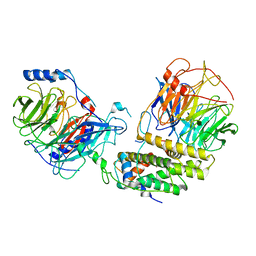 | | Cryo-EM structure of the monomeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IQF
 
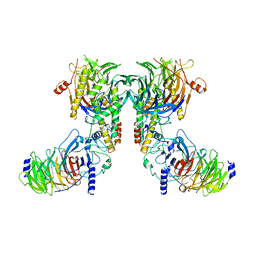 | | Cryo-EM structure of the dimeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
6K15
 
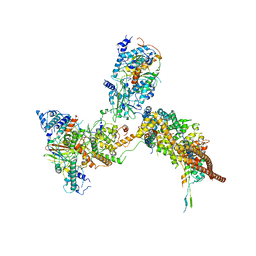 | | RSC substrate-recruitment module | | Descriptor: | Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
5X80
 
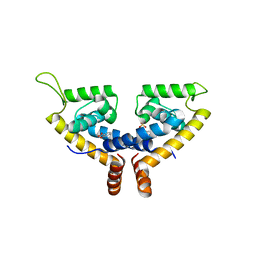 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS MARR FAMILY PROTEIN RV2887 COMPLEX WITH SALICYLIC ACID | | Descriptor: | 2-HYDROXYBENZOIC ACID, SULFATE ION, Uncharacterized HTH-type transcriptional regulator Rv2887 | | Authors: | Gao, Y.R, Li, D.F, Wang, D.C, Bi, L.J. | | Deposit date: | 2017-02-28 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis
Sci Rep, 7, 2017
|
|
3BPX
 
 | | Crystal Structure of MarR | | Descriptor: | 2-HYDROXYBENZOIC ACID, SODIUM ION, Transcriptional regulator | | Authors: | Saridakis, V, Shahinas, D, Xu, X, Christendat, D. | | Deposit date: | 2007-12-19 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum.
J.Mol.Biol., 377, 2008
|
|
4NC7
 
 | | N-terminal domain of delta-subunit of RNA polymerase complexed with I3C and nickel ions | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, DNA-directed RNA polymerase subunit delta, NICKEL (II) ION | | Authors: | Demo, G, Papouskova, V, Komarek, J, Sanderova, H, Rabatinova, A, Krasny, L, Zidek, L, Sklenar, V, Wimmerova, M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray vs. NMR structure of N-terminal domain of delta-subunit of RNA polymerase.
J.Struct.Biol., 187, 2014
|
|
3BPV
 
 | | Crystal Structure of MarR | | Descriptor: | Transcriptional regulator | | Authors: | Saridakis, V, Shahinas, D, Xu, X, Christendat, D. | | Deposit date: | 2007-12-19 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum.
J.Mol.Biol., 377, 2008
|
|
4ETS
 
 | |
6RLB
 
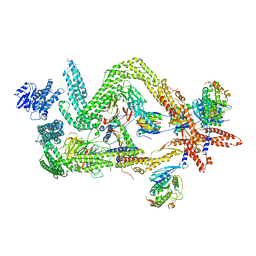 | | Structure of the dynein-2 complex; tail domain | | Descriptor: | Cytoplasmic dynein 2 light intermediate chain 1, Dynein light chain 1, cytoplasmic, ... | | Authors: | Toropova, K, Zalyte, R, Mukhopadhyay, A.G, Mladenov, M, Carter, A.P, Roberts, A.J. | | Deposit date: | 2019-05-01 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the dynein-2 complex and its assembly with intraflagellar transport trains.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8SXU
 
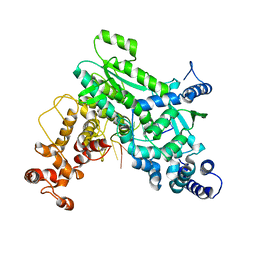 | |
5M4S
 
 | | Transcription factor TFIIA as a single chain protein | | Descriptor: | Transcription initiation factor IIA subunit 2,Transcription initiation factor IIA subunit 1,Transcription initiation factor IIA subunit 1 | | Authors: | Kandiah, E, Gupta, K. | | Deposit date: | 2016-10-19 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Architecture of TAF11/TAF13/TBP complex suggests novel regulation properties of general transcription factor TFIID.
Elife, 6, 2017
|
|
3FMS
 
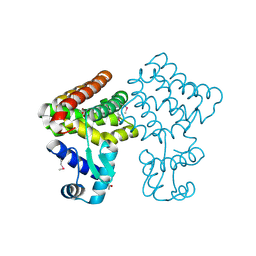 | | Crystal structure of TM0439, a GntR transcriptional regulator | | Descriptor: | ACETATE ION, NICKEL (II) ION, Transcriptional regulator, ... | | Authors: | Zheng, M, Cooper, D.R, Yu, M, Hung, L.-W, Derewenda, U, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2008-12-22 | | Release date: | 2009-02-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Thermotoga maritima TM0439: implications for the mechanism of bacterial GntR transcription regulators with Zn2+-binding FCD domains.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6P0F
 
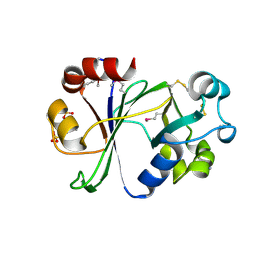 | | N-terminal domain of Thermococcus Gammatolerans McrB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AMMONIUM ION, GTPase subunit of restriction endonuclease, ... | | Authors: | Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-05-17 | | Release date: | 2019-12-18 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | The structure of theThermococcus gammatoleransMcrB N-terminal domain reveals a new mode of substrate recognition and specificity among McrB homologs.
J.Biol.Chem., 295, 2020
|
|
3VOD
 
 | | Crystal Structure of mutant MarR C80S from E.coli | | Descriptor: | Multiple antibiotic resistance protein marR | | Authors: | Lou, H, Zhu, R, Hao, Z. | | Deposit date: | 2012-01-21 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli.
Nat.Chem.Biol., 10, 2014
|
|
2J4D
 
 | | Cryptochrome 3 from Arabidopsis thaliana | | Descriptor: | 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, CRYPTOCHROME DASH, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Klar, T, Pokorny, R, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-28 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryptochrome 3 from Arabidopsis Thaliana: Structural and Functional Analysis of its Complex with a Folate Light Antenna
J.Mol.Biol., 366, 2007
|
|
