1HSS
 
 | | 0.19 ALPHA-AMYLASE INHIBITOR FROM WHEAT | | Descriptor: | 0.19 ALPHA-AMYLASE INHIBITOR | | Authors: | Oda, Y, Fukuyama, K. | | Deposit date: | 1997-07-01 | | Release date: | 1998-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Tertiary and quaternary structures of 0.19 alpha-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 A resolution.
Biochemistry, 36, 1997
|
|
6JDX
 
 | | Crystal structure of AcrIIC2 dimer in complex with partial Nme1Cas9 preprocessed with protease alpha-Chymotrypsin | | Descriptor: | 1,2-ETHANEDIOL, AcrIIC2, CRISPR-associated endonuclease Cas9 | | Authors: | Cheng, Z, Huang, X, Sun, W, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
7RBW
 
 | | Structure of Biliverdin-binding Serpin of Boana punctata (polka-dot tree frog) | | Descriptor: | BILIVERDINE IX ALPHA, Biliverdin bindin serpin | | Authors: | Fedorov, E, Manoilov, K.Y, Verkhusha, V, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of a Biliverdin-Binding Near-Infrared Fluorescent Protein From the Serpin Superfamily.
J.Mol.Biol., 434, 2021
|
|
8WTB
 
 | |
4DKS
 
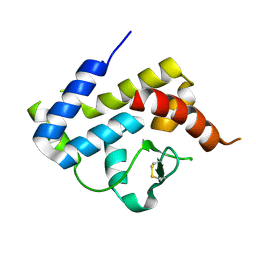 | | A spindle-shaped virus protein (chymotrypsin treated) | | Descriptor: | Probable integrase | | Authors: | Ouyang, S, Liang, W, Huang, L, Liu, Z.-J. | | Deposit date: | 2012-02-04 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of the C-terminal catalytic domain of SSV1 integrase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4K1S
 
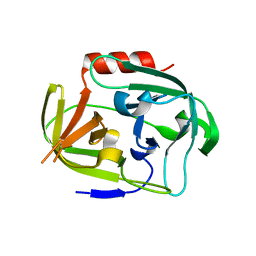 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.96 A resolution | | Descriptor: | Serine protease SplB | | Authors: | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
4K1T
 
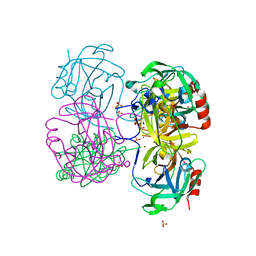 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.60 A resolution | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine protease SplB, ... | | Authors: | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
3W95
 
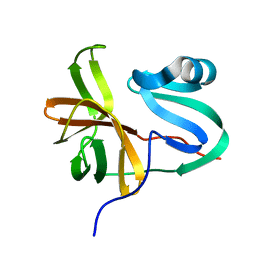 | |
2J92
 
 | |
3R6Y
 
 | | Crystal structure of chymotrypsin-treated aspartase from Bacillus sp. YM55-1 | | Descriptor: | Aspartase, CALCIUM ION | | Authors: | Fibriansah, G, Puthan Veetil, V, Poelarends, G.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry, 50, 2011
|
|
3SFM
 
 | |
1EXF
 
 | | EXFOLIATIVE TOXIN A | | Descriptor: | EXFOLIATVE TOXIN A, GLYCINE | | Authors: | Vath, G.M, Earhart, C.A, Rago, J.V, Kim, M.H, Bohach, G.A, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-10-22 | | Release date: | 1998-02-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the superantigen exfoliative toxin A suggests a novel regulation as a serine protease.
Biochemistry, 36, 1997
|
|
2FYQ
 
 | |
2SEC
 
 | |
2FYR
 
 | |
1WYK
 
 | | SINDBIS VIRUS CAPSID PROTEIN (114-264) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FORMYL GROUP, SINDBIS VIRUS CAPSID PROTEIN | | Authors: | Lee, S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 1998-01-12 | | Release date: | 1998-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the potential glycoprotein binding site of sindbis virus capsid protein with dioxane and model building.
Proteins, 33, 1998
|
|
3WOO
 
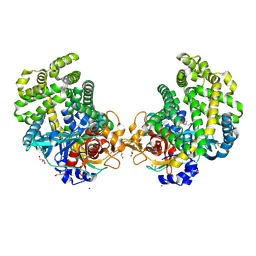 | | Crystal structure of the DAP BII hexapeptide complex I | | Descriptor: | Angiotensin II, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOI
 
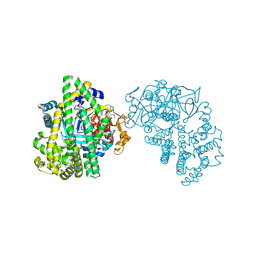 | | Crystal structure of the DAP BII (S657A) | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOK
 
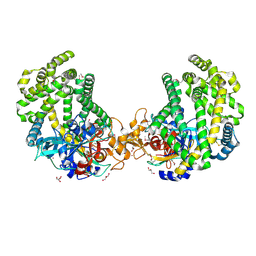 | | Crystal structure of the DAP BII (Space) | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOJ
 
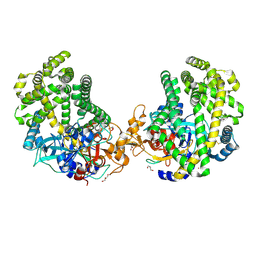 | | Crystal structure of the DAP BII | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
2BHG
 
 | |
3OSY
 
 | |
1ATT
 
 | |
1DS3
 
 | | CRYSTAL STRUCTURE OF OMTKY3-CH2-ASP19I | | Descriptor: | OVOMUCOID | | Authors: | Bateman, K.S, Huang, K, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2000-01-06 | | Release date: | 2001-01-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
3WOP
 
 | | Crystal structure of the DAP BII hexapeptide complex II | | Descriptor: | Angiotensin IV, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
