2II5
 
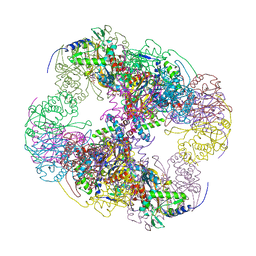 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Isobutyryl-Coenzyme A-bound form | | Descriptor: | ACETATE ION, CHLORIDE ION, ISOBUTYRYL-COENZYME A, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
2II8
 
 | | Anabaena sensory rhodopsin transducer | | Descriptor: | Anabaena sensory rhodopsin transducer protein | | Authors: | Vogeley, L, Sineshchekov, O.A, Trivedi, V.D, Spudich, E.N, Spudich, J.L, Luecke, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the anabaena sensory rhodopsin transducer.
J.Mol.Biol., 367, 2007
|
|
2IKU
 
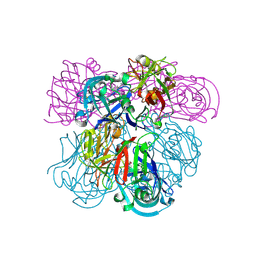 | | Crystal Structure of Human Renin Complexed with Inhibitors | | Descriptor: | 6-ETHYL-5-[(2S)-1-(3-METHOXYPROPYL)-2-PHENYL-1,2,3,4-TETRAHYDROQUINOLIN-7-YL]PYRIMIDINE-2,4-DIAMINE, Renin | | Authors: | Mochalkin, I. | | Deposit date: | 2006-10-02 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding thermodynamics of substituted diaminopyrimidine renin inhibitors.
Anal.Biochem., 360, 2007
|
|
8U1S
 
 | |
8U1C
 
 | |
8U1Q
 
 | |
2II4
 
 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Coenzyme A-bound form | | Descriptor: | CHLORIDE ION, COENZYME A, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
2ILP
 
 | |
2II3
 
 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Oxidized Coenzyme A-bound form | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
7N8R
 
 | | FGTGFG segment from the Nucleoporin p54, residues 63-68 | | Descriptor: | FGTGFG segment from the Nucleoporin p54, residues 63-68, trifluoroacetic acid | | Authors: | Hughes, M.P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Extended beta-Strands Contribute to Reversible Amyloid Formation.
Acs Nano, 16, 2022
|
|
2IND
 
 | | Mn(II) Reconstituted Toluene/o-xylene Monooxygenase Hydroxylase X-ray Crystal Structure | | Descriptor: | HEXAETHYLENE GLYCOL, MANGANESE (II) ION, Toluene, ... | | Authors: | McCormick, M.S, Sazinsky, M.H, Condon, K.L, Lippard, S.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structures of manganese(II)-reconstituted and native toluene/o-xylene monooxygenase hydroxylase reveal rotamer shifts in conserved residues and an enhanced view of the protein interior.
J.Am.Chem.Soc., 128, 2006
|
|
2ITF
 
 | | Crystal structure IsdA NEAT domain from Staphylococcus aureus with heme bound | | Descriptor: | Iron-regulated surface determinant protein A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Grigg, J.C, Vermeiren, C.L, Heinrichs, D.E, Murphy, M.E. | | Deposit date: | 2006-10-19 | | Release date: | 2006-12-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Haem recognition by a Staphylococcus aureus NEAT domain.
Mol.Microbiol., 63, 2007
|
|
2ID4
 
 | |
2DBS
 
 | | Crystal structure of a hypothetical protein TTHC002 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TTHC002 | | Authors: | Yoshikawa, S, Arai, R, Kamo-Uchikubo, T, Fusatomi, E, Akasaka, R, Bessho, Y, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a hypothetical protein TTHC002 from Thermus thermophilus HB8
To be Published
|
|
2DBQ
 
 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (I41) | | Descriptor: | GLYCEROL, Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2IDM
 
 | | 2.00 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | ACETATE ION, Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2IDS
 
 | | Structure of M98A mutant of amicyanin, Cu(I) | | Descriptor: | Amicyanin, COPPER (I) ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
7NKW
 
 | | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-02-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector
To Be Published
|
|
2INC
 
 | | Native Toluene/o-xylene Monooxygenase Hydroxylase X-ray Crystal Structure | | Descriptor: | FE (III) ION, HEXAETHYLENE GLYCOL, Toluene, ... | | Authors: | McCormick, M.S, Sazinsky, M.H, Condon, K.L, Lippard, S.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystal structures of manganese(II)-reconstituted and native toluene/o-xylene monooxygenase hydroxylase reveal rotamer shifts in conserved residues and an enhanced view of the protein interior.
J.Am.Chem.Soc., 128, 2006
|
|
2INZ
 
 | | Crystal Structure of Aldose Reductase complexed with 2-Hydroxyphenylacetic Acid | | Descriptor: | (2-HYDROXYPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Pape, E, Carlson, E, Brownlee, J.M. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2IO5
 
 | | Crystal structure of the CIA- histone H3-H4 complex | | Descriptor: | ASF1A protein, Histone H3.1, Histone H4 | | Authors: | Natsume, R, Akai, Y, Horikoshi, M, Senda, T. | | Deposit date: | 2006-10-10 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4.
Nature, 446, 2007
|
|
2IOD
 
 | | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Dihydroflavonol 4-reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Langlois d'Estaintot, B, Granier, T, Gallois, B. | | Deposit date: | 2006-10-10 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site
To be Published
|
|
2HG0
 
 | | Structure of the West Nile Virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein | | Authors: | Nybakken, G.E, Nelson, C.A, Chen, B.R, Diamond, M.S, Fremont, D.H. | | Deposit date: | 2006-06-26 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the West Nile virus envelope glycoprotein.
J.Virol., 80, 2006
|
|
2HHT
 
 | | C:O6-methyl-guanine pair in the polymerase-2 basepair position | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*TP*CP*G)-3', 5'-D(*GP*TP*AP*CP*(6OG)P*AP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HIN
 
 | | Structure of N15 Cro at 1.05 A: an ortholog of lambda Cro with a completely different but equally effective dimerization mechanism | | Descriptor: | Repressor protein, SULFATE ION | | Authors: | Dubrava, M.S, Ingram, W.M, Roberts, S.A, Weichsel, A, Montfort, W.R, Cordes, M.H. | | Deposit date: | 2006-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | N15 Cro and lambda Cro: orthologous DNA-binding domains with completely different but equally effective homodimer interfaces.
Protein Sci., 17, 2008
|
|
