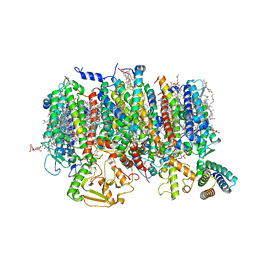
6OVY
 
 | |
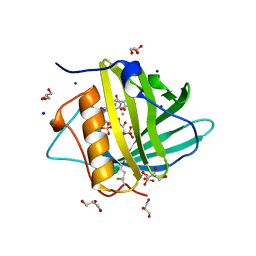
3HWG
 
 | |
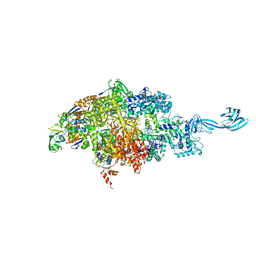
8R2I
 
 | | Cryo-EM Structure of native Photosystem II assembly intermediate from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Fadeeva, M, Klaiman, D, Kandiah, E, Nelson, N. | | Deposit date: | 2023-11-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of native photosystem II assembly intermediate from Chlamydomonas reinhardtii .
Front Plant Sci, 14, 2023
|
|
8QSO
 
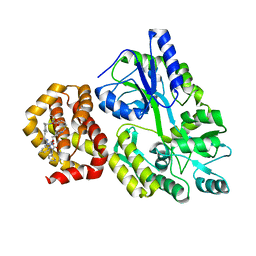 | | Crystal structure of human Mcl-1 in complex with compound 1 | | Descriptor: | (13S,16R,19S)-16-benzyl-43-ethoxy-N-methyl-7,11,14,17-tetraoxo-13-phenyl-5-oxa-2,8,12,15,18-pentaaza-1(1,4),4(1,2)-dibenzena-9(1,4)-cyclohexanacycloicosaphane-19-carboxamide, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hekking, K.F.W, Gremmen, S, Maroto, S, Keefe, A.D, Zhang, Y. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Development of Potent Mcl-1 Inhibitors: Structural Investigations on Macrocycles Originating from a DNA-Encoded Chemical Library Screen.
J.Med.Chem., 67, 2024
|
|
6T5Y
 
 | | Crystal structure of AmpC from E.coli with Zidebactam (WCK 5107) | | Descriptor: | (2S,5R)-1-formyl-N'-[(3R)-piperidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Lang, P.A, Leissing, T.M, Schofield, C.J, Brem, J. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Investigations of the Inhibition of Escherichia coli AmpC beta-Lactamase by Diazabicyclooctanes.
Antimicrob.Agents Chemother., 65, 2021
|
|
5NS2
 
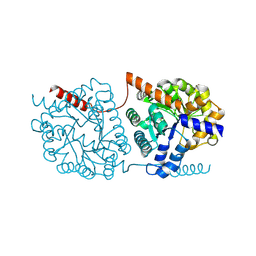 | | Cys-Gly dipeptidase GliJ in complex with Co2+ | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Dipeptidase gliJ, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
2ZDT
 
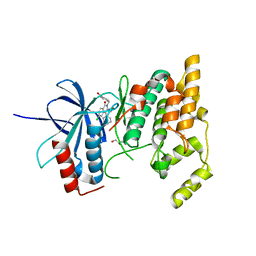 | | Crystal Structure of human JNK3 complexed with an isoquinolone inhibitor | | Descriptor: | 4-[(6-chloro-1-oxo-4-phenyl-3-propanoylisoquinolin-2(1H)-yl)methyl]benzoic acid, GLYCEROL, Mitogen-activated protein kinase 10 | | Authors: | Sogabe, S, Asano, Y, Fukumoto, S, Habuka, N, Fujishima, A. | | Deposit date: | 2007-11-27 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery, synthesis and biological evaluation of isoquinolones as novel and highly selective JNK inhibitors (2)
Bioorg.Med.Chem., 16, 2008
|
|
7TEK
 
 | |
6Y37
 
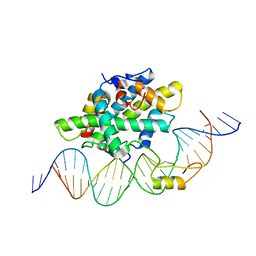 | |
7TEL
 
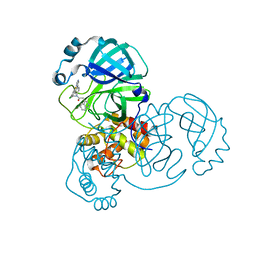 | |
8T0T
 
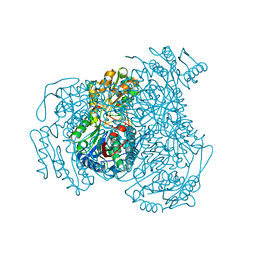 | | Structure of Compound 4 bound to human ALDH1A1 | | Descriptor: | 1-(4-{6-fluoro-3-[4-(methanesulfonyl)piperazine-1-carbonyl]quinolin-4-yl}phenyl)cyclopropane-1-carbonitrile, Aldehyde dehydrogenase 1A1, CHLORIDE ION, ... | | Authors: | Hurley, T.D. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of substituted benzimidazoles as inhibitors of human aldehyde dehydrogenase 1A isoenzymes.
Chem.Biol.Interact., 391, 2024
|
|
3HWE
 
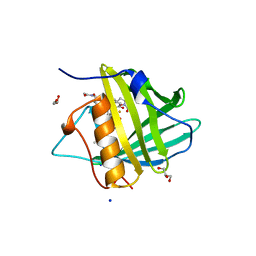 | |
8T0N
 
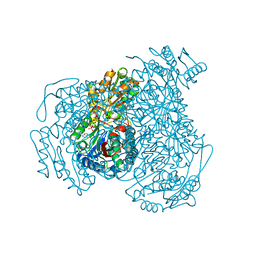 | | Structure of Compound 4 bound to human ALDH1A1 | | Descriptor: | 2-methoxy-6-{[(1-propyl-1H-benzimidazol-2-yl)amino]methyl}phenol, Aldehyde dehydrogenase 1A1, CHLORIDE ION, ... | | Authors: | Hurley, T.D. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of substituted benzimidazoles as inhibitors of human aldehyde dehydrogenase 1A isoenzymes.
Chem.Biol.Interact., 391, 2024
|
|
8COB
 
 | |
6GFG
 
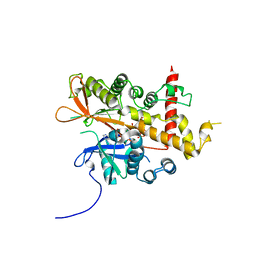 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with D-chiro-IP6 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-chiro inositol hexakisphosphate, Inositol-pentakisphosphate 2-kinase, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
3AR7
 
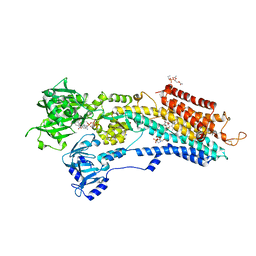 | | Calcium pump crystal structure with bound TNP-ATP and TG in the absence of Ca2+ | | Descriptor: | OCTANOIC ACID [3S-[3ALPHA, 3ABETA, 4ALPHA, ... | | Authors: | Toyoshima, C, Yonekura, S, Tsueda, J, Iwasawa, S. | | Deposit date: | 2010-11-24 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6TFA
 
 | |
3AR5
 
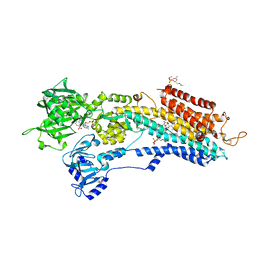 | | Calcium pump crystal structure with bound TNP-AMP and TG | | Descriptor: | 2',3'-O-[(1r)-2,4,6-trinitrocyclohexa-2,5-diene-1,1-diyl]adenosine 5'-(dihydrogen phosphate), OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Toyoshima, C, Yonekura, S, Tsueda, J, Iwasawa, S. | | Deposit date: | 2010-11-24 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6KMX
 
 | | Structure of PSI from H. hongdechloris grown under far-red light condition | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-08-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structural basis for the adaptation and function of chlorophyll f in photosystem I.
Nat Commun, 11, 2020
|
|
7SA3
 
 | | Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Gisriel, C.J, Bryant, D.A, Brudvig, G.W. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light reveals the functions of chlorophylls d and f.
J.Biol.Chem., 298, 2021
|
|
6TPM
 
 | | Crystal structure of AmpC from E.coli with Relebactam (MK-7655) | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Lang, P.A, Leissing, T.M, Schofield, C.J, Brem, J. | | Deposit date: | 2019-12-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Investigations of the Inhibition of Escherichia coli AmpC beta-Lactamase by Diazabicyclooctanes.
Antimicrob.Agents Chemother., 65, 2021
|
|
3K0J
 
 | | Crystal structure of the E. coli ThiM riboswitch in complex with thiamine pyrophosphate and the U1A crystallization module | | Descriptor: | MAGNESIUM ION, RNA (87-MER), THIAMINE DIPHOSPHATE, ... | | Authors: | Kulshina, N, Edwards, T.E, Ferre-D'Amare, A.R. | | Deposit date: | 2009-09-24 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch.
Rna, 16, 2010
|
|
3K3L
 
 | |
5OE2
 
 | | CRYSTAL STRUCTURE OF THE BETA-LACTAMASE OXA-245 | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION | | Authors: | Lund, B.A, Leiros, H.K.S. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6R1P
 
 | | EthR ligand complex | | Descriptor: | 2-[2-[4-(2,3-dihydro-1,4-benzodioxin-6-yl)-1,2,3-triazol-1-yl]ethyl]-6-methyl-1~{H}-pyrimidin-4-one, HTH-type transcriptional regulator EthR | | Authors: | Pohl, E, Tatum, N. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Relative Binding Energies Predict Crystallographic Binding Modes of Ethionamide Booster Lead Compounds.
J Phys Chem Lett, 10, 2019
|
|
