3N5J
 
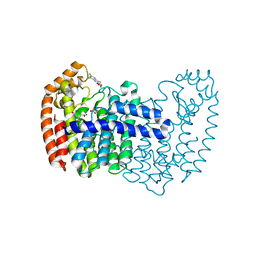 | | Human fpps complex with NOV_311 | | Descriptor: | 3-(carboxymethyl)-5,7-dichloro-1H-indole-2-carboxylic acid, FARNESYL PYROPHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Rondeau, J.-M. | | Deposit date: | 2010-05-25 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
Nat.Chem.Biol., 6, 2010
|
|
8BQY
 
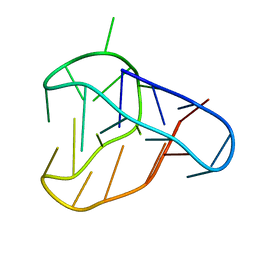 | | An i-motif domain able to undergo pH-dependent conformational transitions (acidic structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*(DNR)P*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*(DNR)P*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
4J97
 
 | |
8BV6
 
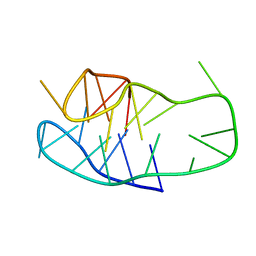 | | An i-motif domain able to undergo pH-dependent conformational transitions (neutral structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8BXA
 
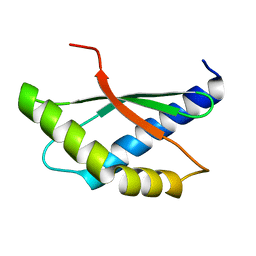 | | Crystal structure of ribosome binding factor A (RbfA) from S. aureus | | Descriptor: | Ribosome-binding factor A | | Authors: | Fatkhullin, B, Bikmullin, A, Gabdulkhakov, A, Khusainov, I, Validov, S, Usachev, K, Yusupov, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Yet Another Similarity between Mitochondrial and Bacterial Ribosomal Small Subunit Biogenesis Obtained by Structural Characterization of RbfA from S. aureus.
Int J Mol Sci, 24, 2023
|
|
3MMN
 
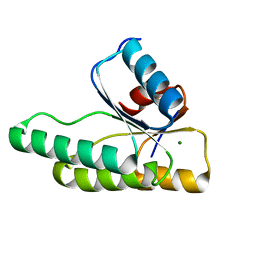 | | Crystal structure of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana complexed with Mg2+ | | Descriptor: | Histidine kinase homolog, MAGNESIUM ION | | Authors: | Marek, J, Klumpler, T, Pekarova, B, Triskova, O, Horak, J, Zidek, L, Dopitova, R, Hejatko, J, Janda, L. | | Deposit date: | 2010-04-20 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and binding specificity of the receiver domain of sensor histidine kinase CKI1 from Arabidopsis thaliana.
Plant J., 67, 2011
|
|
4J96
 
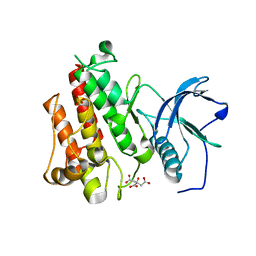 | |
4JNK
 
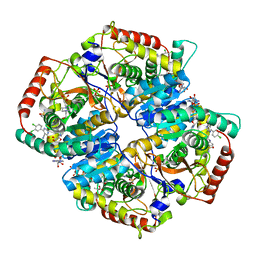 | | Lactate Dehydrogenase A in complex with inhibitor compound 22 | | Descriptor: | (2R)-2-{[5-cyano-4-(3,4-dichlorophenyl)-6-oxo-1,6-dihydropyrimidin-2-yl]sulfanyl}-N-(4-sulfamoylphenyl)propanamide, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Identification of substituted 2-thio-6-oxo-1,6-dihydropyrimidines as inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JVQ
 
 | | Crystal structure of hcv ns5b polymerase in complex with compound 9 | | Descriptor: | 5-{4-[(4-methoxybenzoyl)amino]phenoxy}-2-{[(trans-4-methylcyclohexyl)carbonyl](propan-2-yl)amino}benzoic acid, GLYCEROL, Genome polyprotein, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-26 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enantiomeric Atropisomers Inhibit HCV Polymerase and/or HIV Matrix: Characterizing Hindered Bond Rotations and Target Selectivity.
J.Med.Chem., 57, 2014
|
|
3M5C
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302E mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
4J74
 
 | | The 1.2A crystal structure of humanized Xenopus MDM2 with RO0503918 - a nutlin fragment | | Descriptor: | (4S,5R)-4,5-bis(4-chlorophenyl)-2-methyl-4,5-dihydro-1H-imidazole, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Janson, C, Lukacs, C, Kammlott, U, Graves, B. | | Deposit date: | 2013-02-12 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Deconstruction of a nutlin: dissecting the binding determinants of a potent protein-protein interaction inhibitor.
ACS Med Chem Lett, 4, 2013
|
|
4J7E
 
 | | The 1.63A crystal structure of humanized Xenopus MDM2 with a nutlin fragment, RO5524529 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(4S,5R)-4,5-bis(4-chlorophenyl)-2,4,5-trimethyl-4,5-dihydro-1H-imidazol-1-yl]{4-[3-(methylsulfonyl)propyl]piperazin-1-yl}methanone | | Authors: | Janson, C, Lukacs, C, Graves, B. | | Deposit date: | 2013-02-13 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Deconstruction of a nutlin: dissecting the binding determinants of a potent protein-protein interaction inhibitor.
ACS Med Chem Lett, 4, 2013
|
|
8R1I
 
 | |
3MPN
 
 | | F177R1 mutant of LeuT | | Descriptor: | CHLORIDE ION, LEUCINE, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ... | | Authors: | Kroncke, B.M. | | Deposit date: | 2010-04-27 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural origins of nitroxide side chain dynamics on membrane protein alpha-helical sites.
Biochemistry, 49, 2010
|
|
8BK6
 
 | | A truncated structure of LpMIP with bound inhibitor JK095. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase | | Authors: | Whittaker, J.J, Guskov, A, Hellmich, A.U, Goretzki, B. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BBK
 
 | |
8SNZ
 
 | | X-ray Crystal Structure of FMN-bound long-chain flavodoxin from Rhodopseudomonas palustris | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Ansari, A, Khan, S.A, Miller, A.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure, dynamics, and redox reactivity of an all-purpose flavodoxin.
J.Biol.Chem., 300, 2024
|
|
6WJ9
 
 | | UDP-GlcNAc C4-epimerase mutant S121A/Y146F from Pseudomonas protegens in complex with UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Willams, R.J, Whitney, J.C, Whitfield, G.B, Robinson, H, Parsek, M.R, Nitz, M, Harrison, J.J, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
7ZO5
 
 | | L1 metallo-beta-lactamase in complex with a mecillinam degradation product | | Descriptor: | (2~{R},4~{S})-2-[(1~{R})-2-(azepan-1-yl)-1-formamido-2-oxidanylidene-ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7ZO2
 
 | | L1 metallo-beta-lactamase complex with hydrolysed doripenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-3-methyl-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, CHLORIDE ION, Metallo-beta-lactamase L1, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7ZO3
 
 | | L1 metallo-beta-lactamase in complex with hydrolysed tebipenem | | Descriptor: | (2S,3R,4S)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[1-(4,5-dihydro-1,3-thiazol-2-yl)azetidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
6WTM
 
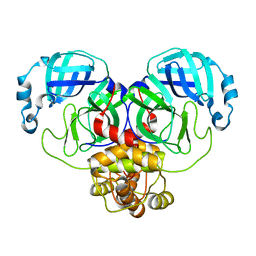 | |
7ZO4
 
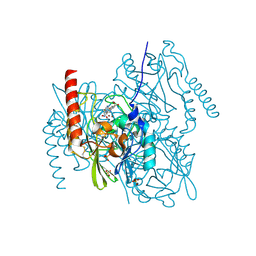 | | L1 metallo-beta-lactamase in complex with hydrolysed panipenem | | Descriptor: | (2R,4S)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S)-1-ethanimidoylpyrrolidin-3-yl]sulfanyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase L1, SODIUM ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7ZO6
 
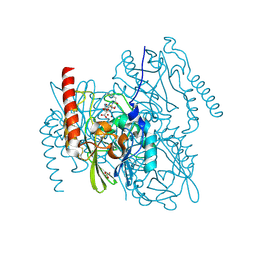 | | L1 metallo-beta-lactamase in complex with hydrolysed cefoxitin | | Descriptor: | (2~{R},5~{S})-5-(aminocarbonyloxymethyl)-2-[(1~{S})-1-methoxy-2-oxidanyl-2-oxidanylidene-1-(2-thiophen-2-ylethanoylamino)ethyl]-5,6-dihydro-2~{H}-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7ZO7
 
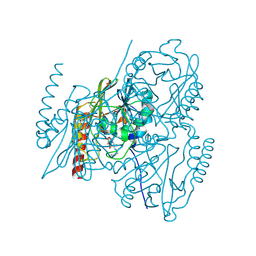 | | L1 metallo-beta-lactamase in complex with hydrolysed cefmetazole | | Descriptor: | (2R,5R)-2-[(1S)-1-[2-(cyanomethylsulfanyl)ethanoylamino]-1-methoxy-2-oxidanyl-2-oxidanylidene-ethyl]-5-[(1-methyl-1,2,3,4-tetrazol-5-yl)sulfanylmethyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
