3BYT
 
 | |
6HZL
 
 | | Crystal structure of redox-inhibited phosphoribulokinase from Synechococcus sp. (strain PCC 6301), osmate derivative | | Descriptor: | OSMIUM ION, Phosphoribulokinase | | Authors: | Wilson, R.H, Bracher, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-27 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of phosphoribulokinase from Synechococcus sp. strain PCC 6301.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3BZY
 
 | | Crystal structure of the mutated Y316D EscU C-terminal domain | | Descriptor: | EscU, SULFATE ION | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
6DXR
 
 | |
3C3C
 
 | | Crystal Structure of human phosphoglycerate kinase bound to 3-phosphoglycerate and L-CDP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Arold, S.T, Gondeau, C, Lionne, C, Chaloin, L. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase
Nucleic Acids Res., 36, 2008
|
|
3C09
 
 | | Crystal structure the Fab fragment of matuzumab (Fab72000) in complex with domain III of the extracellular region of EGFR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Matuzumab Fab Heavy chain, ... | | Authors: | Ferguson, K.M, Schmiedel, J, Knoechel, T. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Matuzumab binding to EGFR prevents the conformational rearrangement required for dimerization.
Cancer Cell, 13, 2008
|
|
7SGP
 
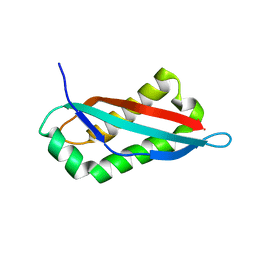 | |
6D6A
 
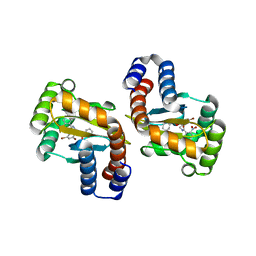 | |
3C16
 
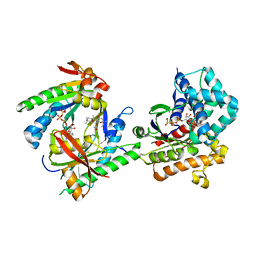 | |
3C1M
 
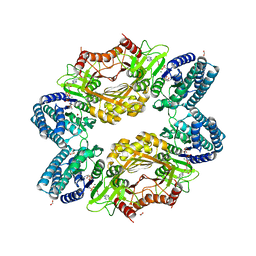 | |
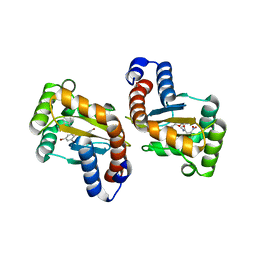
6D6M
 
 | |
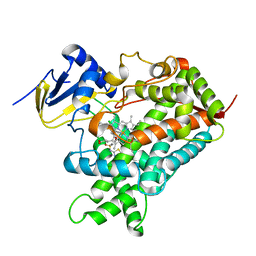
5K7K
 
 | |
8QYI
 
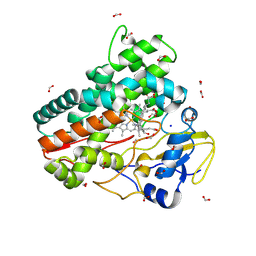 | | OleP in complex with lithocholic acid in high salt crystallization conditions | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Fata, F, Costanzo, A, Freda, I, Gugole, E, Bulfaro, G, Barbizzi, L, Di Renzo, M, Savino, C, Vallone, B, Montemiglio, L.C. | | Deposit date: | 2023-10-26 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | OleP in complex with lithocolic acid in high salt crystallization conditions
To Be Published
|
|
3C6V
 
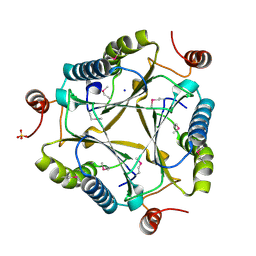 | | Crystal structure of AU4130/APC7354, a probable enzyme from the thermophilic fungus Aspergillus fumigatus | | Descriptor: | CHLORIDE ION, Probable tautomerase/dehalogenase AU4130, SODIUM ION, ... | | Authors: | Singer, A.U, Binkowski, T.A, Skarina, T, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-05 | | Release date: | 2008-02-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of AU4130/APC7354, a probable enzyme from the thermophilic fungus Aspergillus fumigatus.
To be Published
|
|
7SFA
 
 | |
6HW6
 
 | | Yeast 20S proteasome in complex with 20 | | Descriptor: | MAGNESIUM ION, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
3C74
 
 | |
3C32
 
 | |
3C3F
 
 | |
3C3O
 
 | | ALIX Bro1-domain:CHMIP4A co-crystal structure | | Descriptor: | Charged multivesicular body protein 4a peptide, GLYCEROL, Programmed cell death 6-interacting protein | | Authors: | McCullough, J.B, Fisher, R.D, Whitby, F.G, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | ALIX-CHMP4 interactions in the human ESCRT pathway.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CA4
 
 | | Sambucus nigra agglutinin II, tetragonal crystal form- complexed to lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3CAN
 
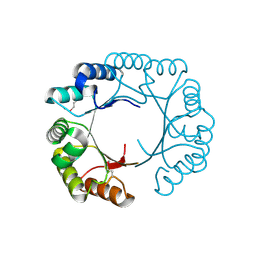 | | Crystal structure of a domain of pyruvate-formate lyase-activating enzyme from Bacteroides vulgatus ATCC 8482 | | Descriptor: | Pyruvate-formate lyase-activating enzyme | | Authors: | Nocek, B, Hendricks, R, Hatzos, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a domain of pyruvate-formate lyase-activating enzyme from Bacteroides vulgatus ATCC 8482.
To be Published
|
|
8RFQ
 
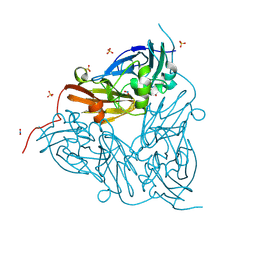 | | Low pH (5.5) nitrite-bound MSOX movie series dataset 5 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [3.05 MGy] - nitric oxide (NO) intermediate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
3C4B
 
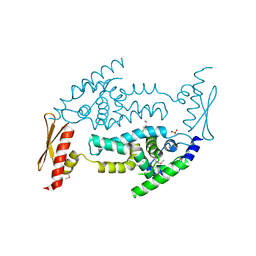 | | Structure of RNaseIIIb and dsRNA binding domains of mouse Dicer | | Descriptor: | Endoribonuclease Dicer | | Authors: | Lee, J.K, Du, Z, Tjhen, R.J, Stroud, R.M, James, T.L. | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and biochemical insights into the dicing mechanism of mouse Dicer: A conserved lysine is critical for dsRNA cleavage.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5JVW
 
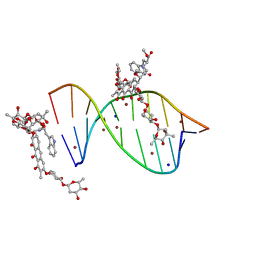 | | Crystal structure of mithramycin analogue MTM SA-Trp in complex with a 10-mer DNA AGAGGCCTCT. | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*GP*CP*CP*TP*CP*T)-3'), Plicamycin, mithramycin analogue MTM SA-Trp, ... | | Authors: | Hou, C, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2016-05-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1.
Nucleic Acids Res., 44, 2016
|
|
