2MFX
 
 | | Non-reducible analogues of alpha-conotoxin Vc1.1: [2,8]-cis dicarba Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Robinson, S.D, Macraild, C.A, Van Lierop, B.J, Robinson, A.J, Norton, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2013-12-18 | | Method: | SOLUTION NMR | | Cite: | Dicarba alpha-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors.
Acs Chem.Biol., 8, 2013
|
|
2M2P
 
 | |
2MGT
 
 | |
2MCU
 
 | | Solid-state NMR structure of piscidin 1 in aligned 3:1 phosphatidylcholine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2MIC
 
 | |
2MIT
 
 | |
2ME7
 
 | |
2MMT
 
 | | Lasso peptide-based integrin inhibitor: Microcin J25 variant with RGDF substitution of Gly12-Ile13-Gly14-Thr15 | | Descriptor: | Microcin J25 RGDF mutant | | Authors: | Hegemann, J.D, Zimmermann, M, Knappe, T.A, Xie, X, Marahiel, M.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-07-02 | | Last modified: | 2014-07-23 | | Method: | SOLUTION NMR | | Cite: | Rational improvement of the affinity and selectivity of integrin binding of grafted lasso peptides.
J.Med.Chem., 57, 2014
|
|
2M0Z
 
 | | cis form of a photoswitchable PDZ domain crosslinked with an azobenzene derivative | | Descriptor: | 3,3'-(E)-diazene-1,2-diylbis{6-[(chloroacetyl)amino]benzenesulfonic acid}, Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Walser, R, Zerbe, O, Hamm, P. | | Deposit date: | 2012-11-09 | | Release date: | 2013-07-03 | | Last modified: | 2013-08-07 | | Method: | SOLUTION NMR | | Cite: | Kinetic response of a photoperturbed allosteric protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2M96
 
 | |
2MLA
 
 | |
2MEO
 
 | |
2LYZ
 
 | |
2MHA
 
 | | CRYSTAL STRUCTURE OF THE MAJOR HISTOCOMPATIBILITY COMPLEX CLASS I H-2KB MOLECULE CONTAINING A SINGLE VIRAL PEPTIDE: IMPLICATIONS FOR PEPTIDE BINDING AND T-CELL RECEPTOR RECOGNITION | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (H-2KB) (ALPHA CHAIN), VIRAL OCTAPEPTIDE ARG-GLY-TYR-VAL-TYR-GLN-GLY-LEU | | Authors: | Zhang, W, Young, A.C.M, Imarai, M, Nathenson, S.G, Sacchettini, J.C. | | Deposit date: | 1993-07-21 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the major histocompatibility complex class I H-2Kb molecule containing a single viral peptide: implications for peptide binding and T-cell receptor recognition.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
2M2M
 
 | |
2M61
 
 | | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails | | Descriptor: | Conotoxin Ar1446 | | Authors: | Sarma, S.P, Rajesh, R.P, Kumar, G.S, Sudarslal, S, Sabareesh, V, Gowd, K.H, Gupta, K, Krishnan, K.S, Balaram, P. | | Deposit date: | 2013-03-18 | | Release date: | 2014-04-16 | | Method: | SOLUTION NMR | | Cite: | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails
To be Published
|
|
2LZS
 
 | | TatA oligomer | | Descriptor: | Sec-independent protein translocase protein TatA | | Authors: | Rodriguez, F.M, Berks, B.C, Schnell, J.R. | | Deposit date: | 2012-10-08 | | Release date: | 2013-03-20 | | Last modified: | 2013-04-24 | | Method: | SOLUTION NMR | | Cite: | Structural model for the protein-translocating element of the twin-arginine transport system.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2MCY
 
 | | CR1 Sushi domains 2 and 3 | | Descriptor: | Complement receptor type 1 | | Authors: | Park, H.J, Guariento, M.J, Maciejewski, M, Hauart, R, Tham, W, Cowman, A.F, Schmidt, C.Q, Martens, H, Liszewski, K.M, Hourcade, D, Barlow, P.N, Atkinson, J.P. | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2014-01-22 | | Method: | SOLUTION NMR | | Cite: | Using Mutagenesis and Structural Biology to Map the Binding Site for the Plasmodium falciparum Merozoite Protein PfRh4 on the Human Immune Adherence Receptor.
J.Biol.Chem., 289, 2014
|
|
2MLM
 
 | | Solution structure of sortase A from S. aureus in complex with benzo[d]isothiazol-3-one based inhibitor | | Descriptor: | N-{2-oxo-2-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylamino]ethyl}-2-sulfanylbenzamide, Sortase family protein | | Authors: | Jaudzems, K, Zhulenkovs, D, Leonchiks, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-11-19 | | Last modified: | 2018-06-27 | | Method: | SOLUTION NMR | | Cite: | Discovery and structure-activity relationship studies of irreversible benzisothiazolinone-based inhibitors against Staphylococcus aureus sortase A transpeptidase.
Bioorg.Med.Chem., 22, 2014
|
|
2MIZ
 
 | |
8QZE
 
 | | Heme-domain BM3 variant 21B3_F87V-A328F | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Opperman, D.J, Ebrecht, A.C, Aschenbrenner, J.C. | | Deposit date: | 2023-10-27 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Revisiting strategies and their combinatorial effect for introducing peroxygenase activity in CYP102A1 (P450BM3)
Mol Catal, 557, 2024
|
|
8QZF
 
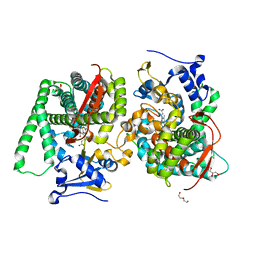 | | Heme-domain BM3 mutant T268E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Opperman, D.J, Ebrecht, A.C, Aschenbrenner, J.C. | | Deposit date: | 2023-10-27 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Revisiting strategies and their combinatorial effect for introducing peroxygenase activity in CYP102A1 (P450BM3)
Mol Catal, 557, 2024
|
|
8PS0
 
 | |
6OHE
 
 | |
6ONA
 
 | |
