5G6T
 
 | | Crystal structure of Zn-containing NagZ H174A mutant from Pseudomonas aeruginosa | | Descriptor: | BETA-HEXOSAMINIDASE, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-07-15 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5GIY
 
 | | HSA-Palmitic acid-[RuCl5(ind)]2- | | Descriptor: | PALMITIC ACID, Serum albumin, pentakis(chloranyl)-(1~{H}-indazol-2-ium-2-yl)ruthenium(1-) | | Authors: | Yang, F, Wang, T. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Structure of HSA-Palmitic acid-[RuCl5(ind)]2-
To Be Published
|
|
5G5U
 
 | | Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa | | Descriptor: | BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-06-05 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
6PZM
 
 | |
5GNM
 
 | | Cytochrome P450 Vdh (CYP107BR1) L348M mutant | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Vitamin D(3) 25-hydroxylase | | Authors: | Yasutake, Y, Tamura, T. | | Deposit date: | 2016-07-22 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the mechanism of the drastic changes in enzymatic activity of the cytochrome P450 vitamin D3 hydroxylase (CYP107BR1) caused by a mutation distant from the active site
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5GOR
 
 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 | | Descriptor: | Alkaline Invertase, GLYCEROL, SULFATE ION | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.673 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
6Q63
 
 | | BT0459 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase, CALCIUM ION, ... | | Authors: | Basle, A, Crouch, L, Bolam, D. | | Deposit date: | 2018-12-10 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci.
Nat Microbiol, 4, 2019
|
|
5G27
 
 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at Room Temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, ENDOLYSIN, ... | | Authors: | Gohlke, U, Consentius, P, Loll, B, Mueller, R, Kaupp, M, Heinemann, U, Risse, T. | | Deposit date: | 2016-04-07 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-Ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
5GM4
 
 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose | | Descriptor: | Endoglucanase-1, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
6Q6K
 
 | | Crystal structure of recombinant human beta-glucocerebrosidase in complex with cyclophellitol activity based probe with Cy5 tag (ME569) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[3,3-dimethyl-2-[(1~{E},3~{E},5~{E})-5-(1,3,3-trimethylindol-2-ylidene)penta-1,3-dienyl]indol-1-ium-1-yl]-~{N}-[[1-[[(1~{S},2~{R},3~{R},4~{S},6~{S})-2,3,4,6-tetrakis(oxidanyl)cyclohexyl]methyl]-1,2,3-triazol-4-yl]methyl]hexanamide, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Functionalized Cyclophellitols Are Selective Glucocerebrosidase Inhibitors and Induce a Bona Fide Neuropathic Gaucher Model in Zebrafish.
J.Am.Chem.Soc., 141, 2019
|
|
6QAO
 
 | |
6Q7J
 
 | | GH3 exo-beta-xylosidase (XlnD) in complex with xylobiose aziridine activity based probe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6QC6
 
 | | Ovine respiratory complex I FRC open class 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-21 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
5GQE
 
 | | Crystal structure of michaelis complex of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 | | Descriptor: | Beta-xylanase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Suzuki, R, Fujimoto, Z, Kaneko, S, Kuno, A. | | Deposit date: | 2016-08-07 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Azidolysis by the Formation of Stable Ser-His Catalytic Dyad in a Glycoside Hydrolase Family 10 Xylanase Mutant
J.Appl.Glyosci., 65, 2019
|
|
6Q8R
 
 | |
5IY5
 
 | | Electron transfer complex of cytochrome c and cytochrome c oxidase at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, S, Baba, J, Aoe, S, Shimada, A, Yamashita, E, Tsukihara, T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode
EMBO J., 36, 2017
|
|
1HU0
 
 | | CRYSTAL STRUCTURE OF AN HOGG1-DNA BOROHYDRIDE TRAPPED INTERMEDIATE COMPLEX | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-OXOGUANINE, ... | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2001-01-03 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Product-Assisted Catalysis in base-excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
5HJA
 
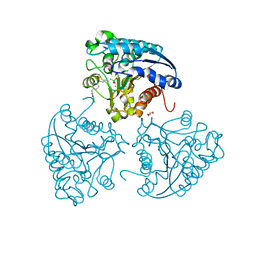 | | Crystal structure of Leishmania mexicana arginase in complex with inhibitor ABHDP | | Descriptor: | (R)-2-amino-6-borono-2-(1-(3,4-dichlorobenzyl)piperidin-4-yl)hexanoic acid, Arginase, GLYCEROL, ... | | Authors: | Hai, Y, Christianson, D.W. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of Leishmania mexicana arginase complexed with alpha , alpha-disubstituted boronic amino-acid inhibitors.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5HJH
 
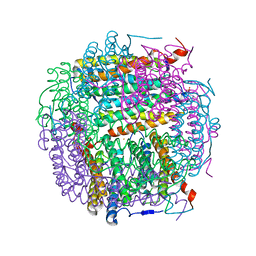 | | Dps4 from Nostoc punctiforme in complex with Fe ions | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FE (III) ION, Ferritin, ... | | Authors: | Howe, C, Moparthi, V.K, Persson, K, Stensjo, K. | | Deposit date: | 2016-01-13 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dps4 from Nostoc punctiforme in complex with Fe ions
To Be Published
|
|
5J31
 
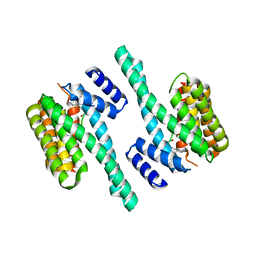 | | Crystal structure of 14-3-3zeta in complex with an alkyne cross-linked cyclic peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, Exoenzyme S | | Authors: | Wallraven, K, Cromm, P, Bier, D, Glas, A, Grossmann, T. | | Deposit date: | 2016-03-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Constraining an Irregular Peptide Secondary Structure through Ring-Closing Alkyne Metathesis.
Chembiochem, 17, 2016
|
|
6QEA
 
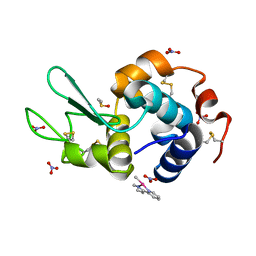 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme and complex I, a pentacoordinate Pt(II) compound containing 2,9-dimethyl-1,10-phenanthroline, dimethylfumarate, methyl and iodine as ligands | | Descriptor: | DIMETHYL SULFOXIDE, Lysozyme C, NITRATE ION, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2019-01-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Reaction with Proteins of a Five-Coordinate Platinum(II) Compound.
Int J Mol Sci, 20, 2019
|
|
5H81
 
 | | HETEROYOHIMBINE SYNTHASE THAS2 FROM CATHARANTHUS ROSEUS - COMPLEX WITH NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION, heteroyohimbine synthase THAS2 | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
6QLJ
 
 | | Crystal structure of F181Q UbiX in complex with an oxidised N5-C1' adduct derived from DMAP | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, Flavin prenyltransferase UbiX, PHOSPHATE ION, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The UbiX flavin prenyltransferase reaction mechanism resembles class I terpene cyclase chemistry.
Nat Commun, 10, 2019
|
|
5H9O
 
 | | Complex of Murine endoplasmic reticulum alpha-glucosidase II with D-Glucose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2015-12-29 | | Release date: | 2016-07-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HY4
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
