8VR2
 
 | | Crystal structure of the Pcryo_0617 oxidoreductase/decarboxylase from Psychrobacter cryohalolentis K5 in the presence of NAD and UDP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NAD-dependent epimerase/dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
8VR3
 
 | | crystal structure of the Pcryo_0618 aminotransferase from Psychrobacter cryohalolentis K5 in the presence of its internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-(1,4-diazepane-1,4-diyl)di(ethane-1-sulfonic acid), CHLORIDE ION, ... | | Authors: | Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
8VR5
 
 | | crystal structure of the Pcryo_0618 aminotransferase from Psychrobacter cryohalolentis K5 in the presence of PMP and glutamate | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-(1,4-diazepane-1,4-diyl)di(ethane-1-sulfonic acid), 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
2A0N
 
 | |
3DB3
 
 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 in complex with trimethylated histone H3-K9 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Trimethylated histone H3-K9 peptide | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-09-16 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
8GSI
 
 | | Structure of the cobolimab Fab | | Descriptor: | heavy chain, light chain, nanobody | | Authors: | Heo, Y.S, Choi, S.B. | | Deposit date: | 2022-09-06 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of the Fab fragment of cobolimab, an investigational antibody targeting Tim-3 for immune-oncology.
To Be Published
|
|
8IFB
 
 | | Dibekacin-bound E.coli 70S ribosome in the PURE system | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
5CQ2
 
 | | Crystal Structure of tandem WW domains of ITCH in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Thioredoxin-interacting protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the regulatory role of the PPxY motifs in the thioredoxin-interacting protein TXNIP.
Biochem.J., 473, 2016
|
|
8VXE
 
 | | Structure of p38 alpha (Mitogen-activated protein kinase 14) complexed with inhibitor 6 | | Descriptor: | (4M)-4-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrrolo[2,3-b]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Blaesse, M, Steinbacher, S, Shaffer, P.L, Sharma, S, Thompson, A.A. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
1UM2
 
 | |
2F46
 
 | |
1Q0M
 
 | | Crystal structure of Ni-containing superoxide dismutase with Ni-ligation corresponding to the state after full x-ray-induced reduction | | Descriptor: | ACETIC ACID, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Wuerges, J, Lee, J.-W, Yim, Y.-I, Yim, H.-S, Kang, S.-O, Djinovic Carugo, K. | | Deposit date: | 2003-07-16 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of nickel-containing superoxide dismutase reveals another type of active site
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q0D
 
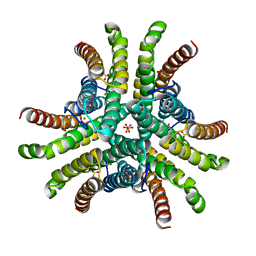 | | Crystal structure of Ni-containing superoxide dismutase with Ni-ligation corresponding to the oxidized state | | Descriptor: | NICKEL (III) ION, SULFATE ION, Superoxide dismutase [Ni] | | Authors: | Wuerges, J, Lee, J.-W, Yim, Y.-I, Yim, H.-S, Kang, S.-O, Djinovic Carugo, K. | | Deposit date: | 2003-07-16 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of nickel-containing superoxide dismutase reveals another type of active site
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1KGI
 
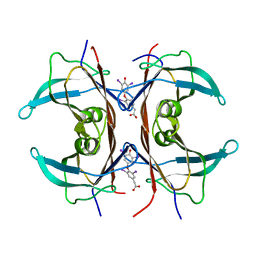 | | Rat transthyretin (also called prealbumin) complex with 3,3',5,5'-tetraiodothyroacetic acid (t4ac) | | Descriptor: | 3,3',5,5'-TETRAIODOTHYROACETIC ACID, TRANSTHYRETIN | | Authors: | Wojtczak, A, Neumann, P, Muziol, T, Cody, V, Luft, J.R, Pangborn, W. | | Deposit date: | 2001-11-27 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex of rat transthyretin with tetraiodothyroacetic acid refined at 2.1 and 1.8 A resolution.
Acta Biochim.Pol., 48, 2001
|
|
2F1L
 
 | |
7NMZ
 
 | |
3G1Q
 
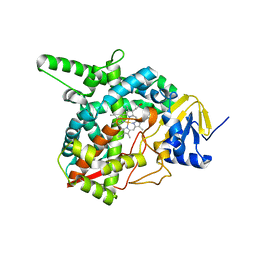 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei in ligand free state | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha-demethylase | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Harp, J, Wawrzak, Z, Waterman, M.R, Park, H. | | Deposit date: | 2009-01-30 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Trypanosoma brucei sterol 14alpha-demethylase and implications for selective treatment of human infections.
J.Biol.Chem., 285, 2010
|
|
2AJ6
 
 | |
1QJS
 
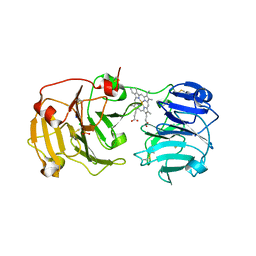 | | mammalian blood serum haemopexin glycosylated-native protein and in complex with its ligand haem | | Descriptor: | CHLORIDE ION, HEMOPEXIN, PHOSPHATE ION, ... | | Authors: | Paoli, M, Baker, H.M, Morgan, W.T, Smith, A, Baker, E.N. | | Deposit date: | 1999-07-01 | | Release date: | 2000-02-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Hemopexin Reveals a Novel High-Affinity Heme Site Formed between Two Beta-Propeller Domains.
Nat.Struct.Biol., 6, 1999
|
|
2OLV
 
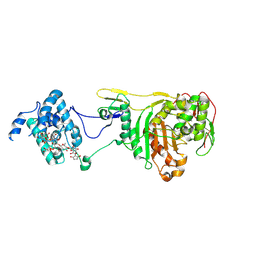 | | Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 2 | | Authors: | Lovering, A.L, De Castro, L, Lim, D, Strynadka, N.C.J. | | Deposit date: | 2007-01-19 | | Release date: | 2007-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis.
Science, 315, 2007
|
|
2K4D
 
 | | E2-c-Cbl recognition is necessary but not sufficient for ubiquitination activity | | Descriptor: | E3 ubiquitin-protein ligase CBL, ZINC ION | | Authors: | Huang, A, De Jong, R.N, Wienk, H, Winkler, S.G, Timmers, H.T.M, Boelens, R. | | Deposit date: | 2008-06-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | E2-c-Cbl recognition is necessary but not sufficient for ubiquitination activity
J.Mol.Biol., 385, 2009
|
|
3PG6
 
 | | The carboxyl terminal domain of human deltex 3-like | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase DTX3L, ... | | Authors: | Walker, J.R, Obiero, J, Kania, J, Schuler, H, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-30 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fold of the conserved DTC domain in Deltex proteins.
Proteins, 80, 2012
|
|
5YY9
 
 | | Crystal structure of Tandem Tudor Domain of human UHRF1 in complex with LIG1-K126me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Ligase 1 | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
3TQX
 
 | | Structure of the 2-amino-3-ketobutyrate coenzyme A ligase (kbl) from Coxiella burnetii | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
1Q0K
 
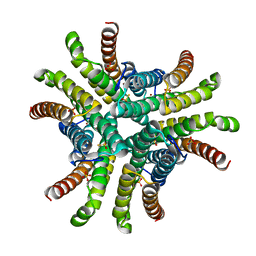 | | Crystal structure of Ni-containing superoxide dismutase with Ni-ligation corresponding to the thiosulfate-reduced state | | Descriptor: | NICKEL (II) ION, SULFATE ION, Superoxide dismutase [Ni], ... | | Authors: | Wuerges, J, Lee, J.-W, Yim, Y.-I, Yim, H.-S, Kang, S.-O, Djinovic Carugo, K. | | Deposit date: | 2003-07-16 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of nickel-containing superoxide dismutase reveals another type of active site
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
