5FWX
 
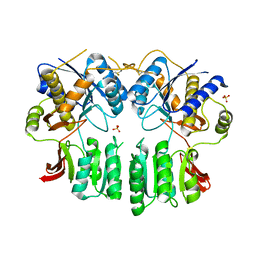 | | Crystal structure of the AMPA receptor GluA2/A4 N-terminal domain heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMATE RECEPTOR 2, GLUTAMATE RECEPTOR 4, ... | | Authors: | Garcia-Nafria, J, Herguedas, B, Greger, I.H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Organization of Heteromeric Ampa-Type Glutamate Receptors.
Science, 352, 2016
|
|
5ET5
 
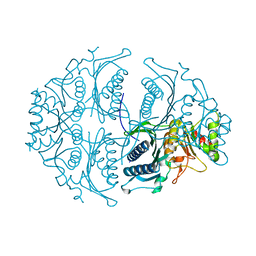 | | Human muscle fructose-1,6-bisphosphatase in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ET8
 
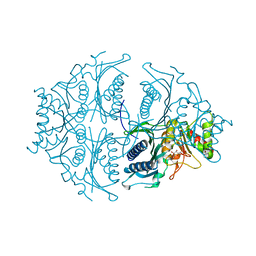 | | Human muscle fructose-1,6-bisphosphatase in active R-state in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | T-to-R switch of muscle FBPase involves extreme changes of secondary and quaternary structure
To Be Published
|
|
6FJG
 
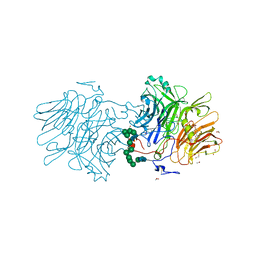 | |
5ET6
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4IPX
 
 | | Analyzing the visible conformational substates of the FK506 binding protein FKBP12 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Chen, H, Mustafi, S.M, Li, H.M, LeMaster, D.M, Hernandez, G. | | Deposit date: | 2013-01-10 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysing the visible conformational substates of the FK506-binding protein FKBP12.
Biochem.J., 453, 2013
|
|
5ET7
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6FDB
 
 | |
1G80
 
 | | NMR SOLUTION STRUCTURE OF D(GCGTACGC)2 | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3' | | Authors: | Isaacs, R.J, Spielmann, H.P. | | Deposit date: | 2000-11-15 | | Release date: | 2001-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
5NB7
 
 | | Complement factor D | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[(6-bromanylpyridin-2-yl)carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]indazole-3-carboxamide, Complement factor D, DIMETHYL SULFOXIDE | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
1BR9
 
 | | HUMAN TISSUE INHIBITOR OF METALLOPROTEINASE-2 | | Descriptor: | METALLOPROTEINASE-2 INHIBITOR | | Authors: | Tuuttila, A, Morgunova, E, Bergmann, U, Lindqvist, Y, Tryggvason, K, Schneider, G. | | Deposit date: | 1998-08-28 | | Release date: | 1999-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of human tissue inhibitor of metalloproteinases-2 at 2.1 A resolution.
J.Mol.Biol., 284, 1998
|
|
5NAR
 
 | |
5NAW
 
 | | Complement factor D in complex with the inhibitor (1R,3S,5R)-2-Aza-bicyclo[3.1.0]hexane-2,3-dicarboxylic acid 2-[(1-carbamoyl-1H-indol-3-yl)-amide] 3-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (1~{R},3~{S},5~{R})-~{N}2-(1-aminocarbonylindol-3-yl)-~{N}3-[3-(trifluoromethyloxy)phenyl]-2-azabicyclo[3.1.0]hexane-2,3-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-02-28 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NBA
 
 | | Complement factor D in complex with the inhibitor (2S,4R)-4-Fluoro-pyrrolidine-1,2-dicarboxylic acid 1-[(1-carbamoyl-1H-indol-3-yl)-amide] 2-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (2~{S},4~{R})-~{N}1-(1-aminocarbonylindol-3-yl)-4-fluoranyl-~{N}2-[3-(trifluoromethyloxy)phenyl]pyrrolidine-1,2-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
6G6W
 
 | | HUMAN PI3KDELTA IN COMPLEX WITH LIGAND LASW1976 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[3-[4-[[(1~{S})-1-(5-methyl-4-oxidanylidene-3-phenyl-pyrrolo[2,1-f][1,2,4]triazin-2-yl)ethyl]amino]-7~{H}-pyrrolo[ 2,3-d]pyrimidin-5-yl]-5-oxidanyl-phenyl]methanesulfonamide | | Authors: | Segarra, V, Hernandez, B, Jestel, A, Mortel, M, Nagel, S. | | Deposit date: | 2018-04-03 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Discovery of a Novel Inhaled PI3K delta Inhibitor for the Treatment of Respiratory Diseases.
J. Med. Chem., 61, 2018
|
|
3JWE
 
 | | Crystal structure of human mono-glyceride lipase in complex with SAR629 | | Descriptor: | 1-[bis(4-fluorophenyl)methyl]-4-(1H-1,2,4-triazol-1-ylcarbonyl)piperazine, MGLL protein | | Authors: | Bertrand, T, Auge, F, Houtmann, J, Rak, A, Vallee, F, Mikol, V, Berne, P.F, Michot, N, Cheuret, D, Hoornaert, C, Mathieu, M. | | Deposit date: | 2009-09-18 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for human monoglyceride lipase inhibition.
J.Mol.Biol., 396, 2010
|
|
1G7Y
 
 | | THE CRYSTAL STRUCTURE OF THE 58KD VEGETATIVE LECTIN FROM THE TROPICAL LEGUME DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, STEM/LEAF LECTIN DB58, ... | | Authors: | Buts, L, Hamelryck, T.W, Loris, R, Dao-Thi, M.-H, Wyns, L, Etzler, M.E. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
3JW8
 
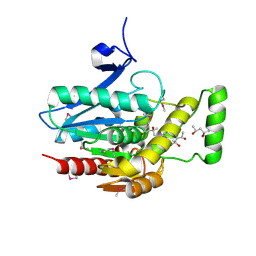 | | Crystal structure of human mono-glyceride lipase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, MGLL protein | | Authors: | Bertrand, T, Auge, F, Houtmann, J, Rak, A, Vallee, F, Mikol, V, Berne, P.F, Michot, N, Cheuret, D, Hoornaert, C, Mathieu, M. | | Deposit date: | 2009-09-18 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for human monoglyceride lipase inhibition.
J.Mol.Biol., 396, 2010
|
|
8BDZ
 
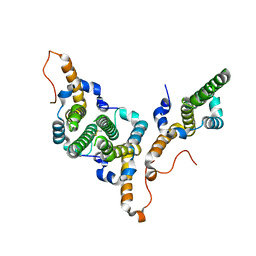 | | Hepatitis B virus core antigen (HBc) with the insertion of four external domains of the influenza A M2 protein (HBc/4M2e) with T=4 topology | | Descriptor: | Core protein,Matrix protein 2,External core antigen | | Authors: | Egorov, V.V, Shvetsov, A.V, Pichkur, E.B, Shaldzhyan, A.A, Zabrodskaya, Y.A, Vinogradova, D.S, Nekrasov, P.A, Gorshkov, A.N, Garmay, Y.P, Kovaleva, A.A, Stepanova, L.A, Tsybalova, L.M, Shtam, T.A, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2022-10-20 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Inside and outside of virus-like particles HBc and HBc/4M2e: A comprehensive study of the structure.
Biophys.Chem., 293, 2022
|
|
5FWY
 
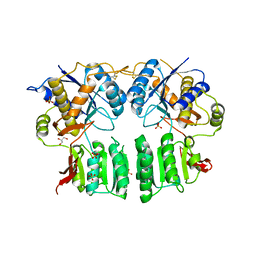 | | Crystal structure of the AMPA receptor GluA2/A3 N-terminal domain heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMATE RECEPTOR 2, GLUTAMATE RECEPTOR 3, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I.H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and Organization of Heteromeric Ampa-Type Glutamate Receptors.
Science, 352, 2016
|
|
8BER
 
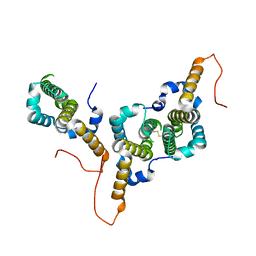 | | Hepatitis B virus core antigen (HBc) with the insertion of four external domains of the influenza A M2 protein (HBc/4M2e) with T=3 topology | | Descriptor: | Core protein,Matrix protein 2,External core antigen | | Authors: | Egorov, V.V, Shvetsov, A.V, Pichkur, E.B, Shaldzhyan, A.A, Zabrodskaya, Y.A, Vinogradova, D.S, Nekrasov, P.A, Gorshkov, A.N, Garmay, Y.P, Kovaleva, A.A, Stepanova, L.A, Tsybalova, L.M, Shtam, T.A, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2022-10-21 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Inside and outside of virus-like particles HBc and HBc/4M2e: A comprehensive study of the structure.
Biophys.Chem., 293, 2022
|
|
5FAH
 
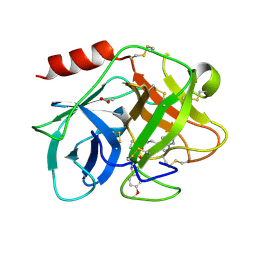 | | KALLIKREIN-7 IN COMPLEX WITH COMPOUND1 | | Descriptor: | (2~{S})-~{N}2-[2-(4-methoxyphenyl)ethyl]-~{N}1-(naphthalen-1-ylmethyl)pyrrolidine-1,2-dicarboxamide, ACETATE ION, Kallikrein-7 | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
1G07
 
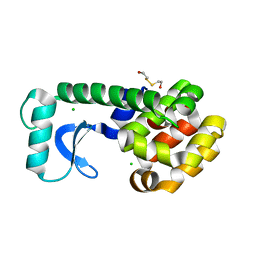 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149C | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-05 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1G0P
 
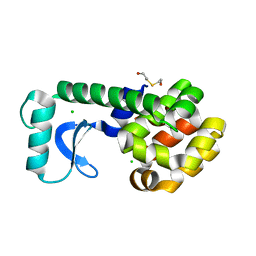 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149G | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1RQ8
 
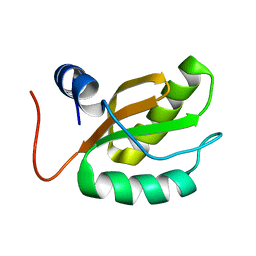 | |
