8FL7
 
 | |
8FLB
 
 | |
7QCM
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3-methoxy-4-hydroxy-acetophenone)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3-metoxy-4-hydroxy-acetophenone)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
6XTX
 
 | | CryoEM structure of human CMG bound to ATPgammaS and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45 homolog, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Rzechorzek, N.J, Pellegrini, L. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | CryoEM structures of human CMG-ATP gamma S-DNA and CMG-AND-1 complexes.
Nucleic Acids Res., 48, 2020
|
|
7QCK
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,5-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,5-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
6Z18
 
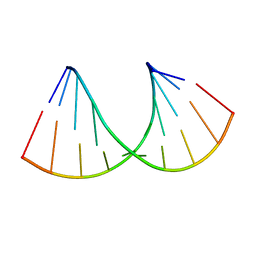 | | Crystal structure of RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG; R32 form | | Descriptor: | RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG | | Authors: | Ruszkowski, M, Sekula, B, Mao, S, Haruehanroengra, P, Sheng, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Base pairing, structural and functional insights into N4-methylcytidine (m4C) and N4,N4-dimethylcytidine (m42C) modified RNA.
Nucleic Acids Res., 48, 2020
|
|
8BNX
 
 | |
6ZD8
 
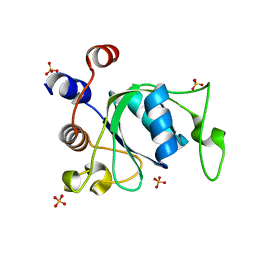 | | Crystal structure of YTHDC1 T379V mutant | | Descriptor: | SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
7LKM
 
 | |
7LKQ
 
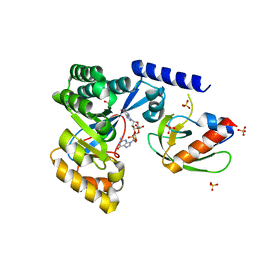 | | The PilZ(delta107-117)-FimX(GGDEF-EAL) complex from Xanthomonas citri | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FimX(GGDEF-EAL), MAGNESIUM ION, ... | | Authors: | Llontop, E.E, Guzzo, C.R, Farah, C.S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The PilB-PilZ-FimX regulatory complex of the Type IV pilus from Xanthomonas citri.
Plos Pathog., 17, 2021
|
|
1JLJ
 
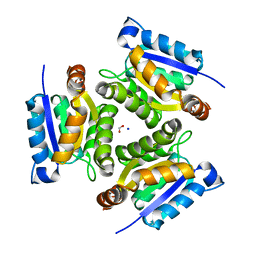 | | 1.6 Angstrom crystal structure of the human neuroreceptor anchoring and molybdenum cofactor biosynthesis protein gephyrin | | Descriptor: | FORMIC ACID, SODIUM ION, gephyrin | | Authors: | Schwarz, G, Schrader, N, Mendel, R.R, Hecht, H.-J, Schindelin, H. | | Deposit date: | 2001-07-16 | | Release date: | 2001-09-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human gephyrin and plant Cnx1 G domains: comparative analysis and functional implications.
J.Mol.Biol., 312, 2001
|
|
7LKO
 
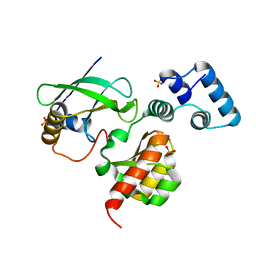 | |
7R3F
 
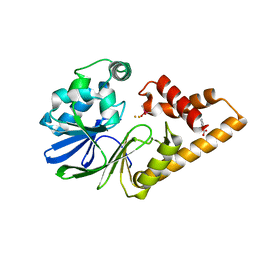 | | Monomeric PqsE mutant E187R | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, BENZOIC ACID, CACODYLATE ION, ... | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7LKN
 
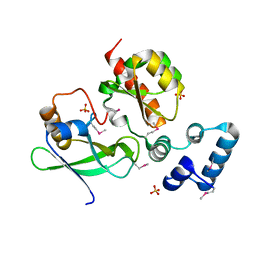 | |
8BNS
 
 | |
7L7P
 
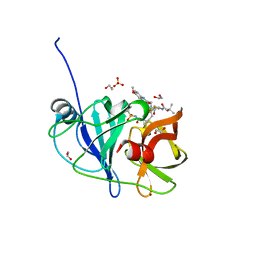 | |
7L7O
 
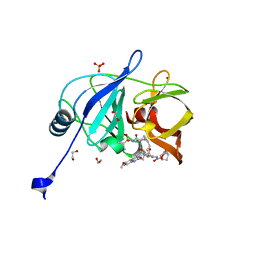 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR04-49 | | Descriptor: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
8BP8
 
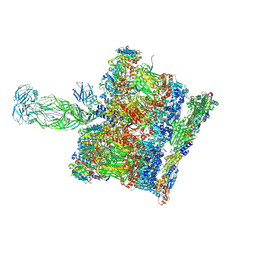 | | SPA of Trypsin untreated Rotavirus TLP spike | | Descriptor: | CALCIUM ION, Inner capsid protein VP2, Intermediate capsid protein VP6, ... | | Authors: | Shah, P.N.M, Stuart, D.I. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
7L7N
 
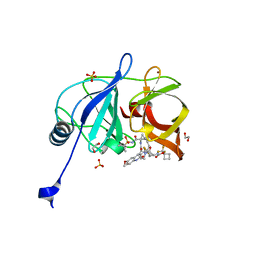 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-59 | | Descriptor: | 1,2-ETHANEDIOL, 1-(trifluoromethyl)cyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
6XTY
 
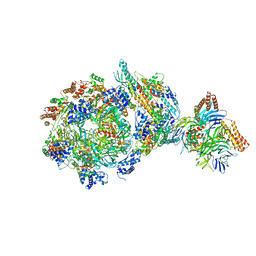 | | CryoEM structure of human CMG bound to AND-1 (CMGA) | | Descriptor: | Cell division control protein 45 homolog, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Rzechorzek, N.J, Pellegrini, L, Chirgadze, D.Y, Hardwick, S.W. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-27 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (6.77 Å) | | Cite: | CryoEM structures of human CMG-ATP gamma S-DNA and CMG-AND-1 complexes.
Nucleic Acids Res., 48, 2020
|
|
7LCA
 
 | |
2CG5
 
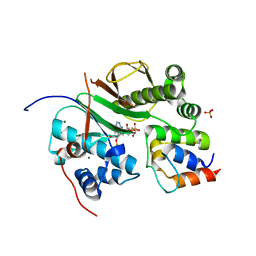 | | Structure of aminoadipate-semialdehyde dehydrogenase- phosphopantetheinyl transferase in complex with cytosolic acyl carrier protein and coenzyme A | | Descriptor: | COENZYME A, FATTY ACID SYNTHASE, L-AMINOADIPATE-SEMIALDEHYDE DEHYDROGENASE-PHOSPHOPANTETHEINYL TRANSFERASE, ... | | Authors: | Bunkoczi, G, Joshi, A, Papagrigoriu, E, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, von Delft, F, Smith, S, Oppermann, U. | | Deposit date: | 2006-02-27 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism and Substrate Recognition of Human Holo Acp Synthase.
Chem.Biol., 14, 2007
|
|
8B37
 
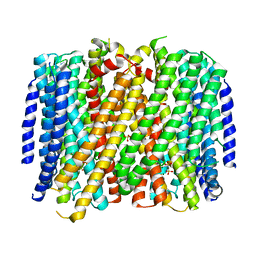 | | Crystal structure of Pyrobaculum aerophilum potassium-independent proton pumping membrane integral pyrophosphatase in complex with imidodiphosphate and magnesium, and with bound sulphate | | Descriptor: | IMIDODIPHOSPHORIC ACID, K(+)-insensitive pyrophosphate-energized proton pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Wilkinson, C, Vidilaseris, K, Ribeiro, O, Liu, J, Hillier, J, Malinen, A, Gehl, B, Jeuken, L.C, Pearson, A.R, Goldman, A. | | Deposit date: | 2022-09-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
6BSW
 
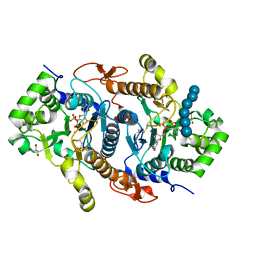 | | Crystal structure of Xyloglucan Xylosyltransferase 1 ternary form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Culbertson, A.T, Ehrlich, J.J, Choe, J, Honzatko, R.B, Zabotina, O.A. | | Deposit date: | 2017-12-04 | | Release date: | 2018-05-23 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6YZZ
 
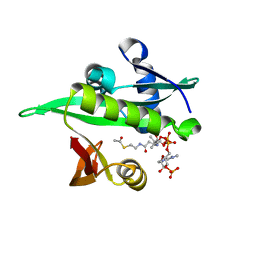 | | Arabidopsis thaliana Naa50 in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
