4NIQ
 
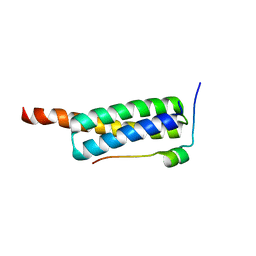 | | Crystal Structure of Vps4 MIT-Vfa1 MIM2 | | Descriptor: | VPS4-associated protein 1, Vacuolar protein sorting-associated protein 4 | | Authors: | Vild, C.J, Xu, Z. | | Deposit date: | 2013-11-06 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Vfa1 Binds to the N-terminal Microtubule-interacting and Trafficking (MIT) Domain of Vps4 and Stimulates Its ATPase Activity.
J.Biol.Chem., 289, 2014
|
|
6EQZ
 
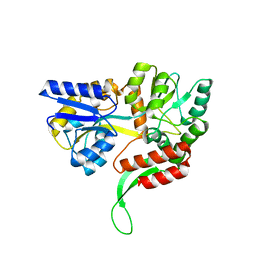 | | A MamC-MIC insertion in MBP scaffold at position K170 | | Descriptor: | Maltose-binding periplasmic protein,Tightly bound bacterial magnetic particle protein,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nudelman, H, Zarivach, R. | | Deposit date: | 2017-10-16 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | The importance of the helical structure of a MamC-derived magnetite-interacting peptide for its function in magnetite formation.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7AZW
 
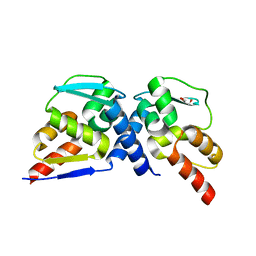 | | Crystal structure of the MIZ1-BTB-domain | | Descriptor: | GLYCEROL, Zinc finger and BTB domain-containing protein 17 isoform X1 | | Authors: | Orth, B, Sander, B, Diederichs, K, Lorenz, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of an atypical interaction site in the BTB domain of the MYC-interacting zinc-finger protein 1.
Structure, 29, 2021
|
|
7AZX
 
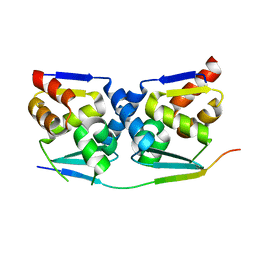 | | Crystal structure of the MIZ1-BTB-domain in complex with a HUWE1-derived peptide | | Descriptor: | E3 ubiquitin-protein ligase HUWE1, Zinc finger and BTB domain-containing protein 17 isoform X1 | | Authors: | Orth, B, Sander, B, Diederichs, K, Lorenz, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of an atypical interaction site in the BTB domain of the MYC-interacting zinc-finger protein 1.
Structure, 29, 2021
|
|
2LSI
 
 | |
1MT3
 
 | | Crystal Structure of the Tricorn Interacting Factor Selenomethionine-F1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-20 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
6RA7
 
 | | Human tnik in complex with compound 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-[8-azanyl-2-(2-fluoranylpyridin-4-yl)-1,7-naphthyridin-5-yl]propan-2-ol, MAGNESIUM ION, ... | | Authors: | Read, J.A. | | Deposit date: | 2019-04-05 | | Release date: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Tool inhibitors and assays to interrogate the biology of the TRAF2 and NCK interacting kinase
To Be Published
|
|
6RA5
 
 | | Human tnik in complex with compound 9 | | Descriptor: | 5-bromanyl-2-(2-fluoranylpyridin-4-yl)-1,7-naphthyridin-8-amine, MAGNESIUM ION, TRAF2 and NCK-interacting protein kinase | | Authors: | Read, J.A. | | Deposit date: | 2019-04-05 | | Release date: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Tool inhibitors and assays to interrogate the biology of the TRAF2 and NCK interacting kinase
To Be Published
|
|
6RO1
 
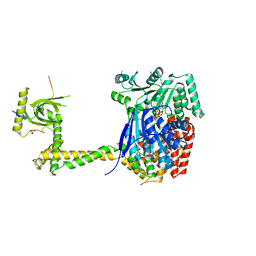 | | X-ray crystal structure of the MTR4 NVL complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Exosome RNA helicase MTR4, ... | | Authors: | Lingaraju, M, Langer, L.M, Basquin, J, Falk, S, Conti, E. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | The MTR4 helicase recruits nuclear adaptors of the human RNA exosome using distinct arch-interacting motifs.
Nat Commun, 10, 2019
|
|
5ZKH
 
 | |
1MTZ
 
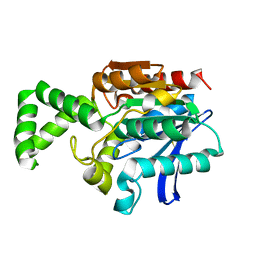 | | Crystal Structure of the Tricorn Interacting Factor F1 | | Descriptor: | Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
1MU0
 
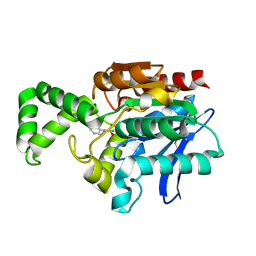 | | Crystal Structure of the Tricorn Interacting Factor F1 Complex with PCK | | Descriptor: | (2R,3S)-3-AMINO-1-CHLORO-4-PHENYL-BUTAN-2-OL, Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
2A25
 
 | | Crystal structure of Siah1 SBD bound to the peptide EKPAAVVAPITTG from SIP | | Descriptor: | Calcyclin-binding protein peptide, Ubiquitin ligase SIAH1, ZINC ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
2YT8
 
 | | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma) | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 3 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Seiki, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma)
To be Published
|
|
7DMB
 
 | |
3BMB
 
 | |
9BJM
 
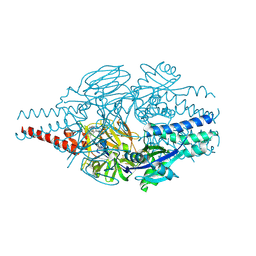 | | Crystal Structure of Inhibitor 5c in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 1'-{[5-chloro-1-(4,4,4-trifluorobutyl)-1H-1,3-benzimidazol-2-yl]methyl}-1-(methanesulfonyl)spiro[azetidine-3,3'-indol]-2'(1'H)-one, Prefusion RSV F (DS-CAV1),Envelope glycoprotein | | Authors: | Shaffer, P.L, Milligan, C, Abeywickrema, P. | | Deposit date: | 2024-04-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Spiro-Azetidine Oxindoles as Long-Acting Injectables for Pre-Exposure Prophylaxis against Respiratory Syncytial Virus Infections.
J.Med.Chem., 67, 2024
|
|
6IY6
 
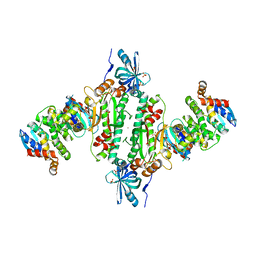 | | Crystal structure of human cytosolic aspartyl-tRNA synthetase (DRS) in complex with glutathion-S transferase (GST) domains from Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 (AIMP2) and glutamyl-prolyl-tRNA synthetase (EPRS) | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, Aspartate--tRNA ligase, cytoplasmic, ... | | Authors: | Park, S.H, Hahn, H, Han, B.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The DRS-AIMP2-EPRS subcomplex acts as a pivot in the multi-tRNA synthetase complex.
Iucrj, 6, 2019
|
|
6IUI
 
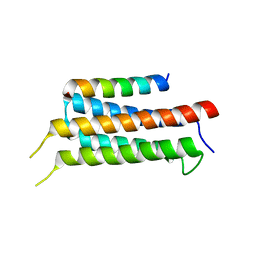 | | Crystal structure of GIT1 PBD domain in complex with Paxillin LD4 motif | | Descriptor: | ARF GTPase-activating protein GIT1, Paxillin | | Authors: | Liang, M, Wei, Z. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the target-binding mode of the G protein-coupled receptor kinase-interacting protein in the regulation of focal adhesion dynamics.
J. Biol. Chem., 294, 2019
|
|
3TIW
 
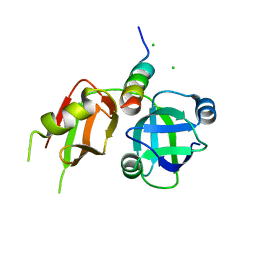 | | Crystal structure of p97N in complex with the C-terminus of gp78 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase AMFR, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2011-08-22 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | The Structural and Functional Basis of the p97/Valosin-containing Protein (VCP)-interacting Motif (VIM): MUTUALLY EXCLUSIVE BINDING OF COFACTORS TO THE N-TERMINAL DOMAIN OF p97.
J.Biol.Chem., 286, 2011
|
|
8QCJ
 
 | |
8QCK
 
 | |
2P4R
 
 | | Structural basis for a novel interaction between AIP4 and beta-PIX | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, GLYCEROL, Rho guanine nucleotide exchange factor 7, ... | | Authors: | Min, K.C. | | Deposit date: | 2007-03-13 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | A novel interaction between atrophin-interacting protein 4 and beta-p21-activated kinase-interactive exchange factor is mediated by an SH3 domain.
J.Biol.Chem., 282, 2007
|
|
3TL8
 
 | |
5XAC
 
 | | CLIR - LC3B | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
