8YE0
 
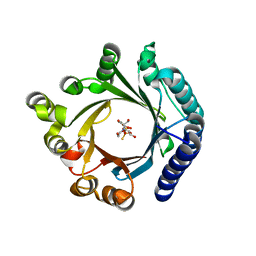 | | Crystal structure of KgpF prenyltransferase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | Authors: | Hamada, K, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2024-02-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | De Novo Discovery of Pseudo-Natural Prenylated Macrocyclic Peptide Ligands.
Angew.Chem.Int.Ed.Engl., 2024
|
|
9BB3
 
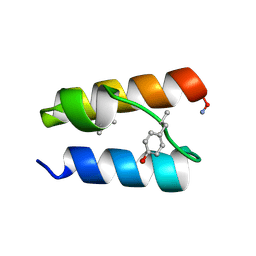 | |
9EO0
 
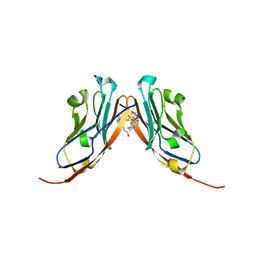 | | Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 | | Descriptor: | Programmed cell death 1 ligand 1, SULFATE ION, ~{N}-[3-[3-[[5-[(2-hydroxyethylamino)methyl]pyridin-2-yl]carbonylamino]-2-methyl-phenyl]-2-methyl-phenyl]-5-[[3-(methylsulfonylamino)propylamino]methyl]pyridine-2-carboxamide | | Authors: | Plewka, J, Hec, A, Sitar, T, Holak, T. | | Deposit date: | 2024-03-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nonsymmetrically Substituted 1,1'-Biphenyl-Based Small Molecule Inhibitors of the PD-1/PD-L1 Interaction.
Acs Med.Chem.Lett., 15, 2024
|
|
9FX5
 
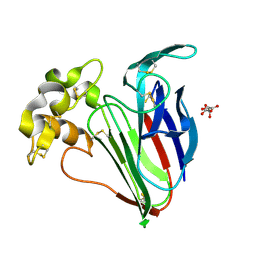 | | Crystal structure of Cryo2RT Thaumatin at 296K | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Huang, C.Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9F9K
 
 | | Crystal structure of MUS81-EME1 bound by compound 15. | | Descriptor: | 5-oxidanyl-6-oxidanylidene-2-(4-phenylphenyl)-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.728 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9BBE
 
 | | Co-crystal structure of human DDB1 bound to fragment UB028668 | | Descriptor: | 5-(4-methoxyphenyl)-3-[(3S)-pyrrolidin-3-yl]-1,2,4-oxadiazole, DNA damage-binding protein 1, L(+)-TARTARIC ACID, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028668
To be published
|
|
8XAI
 
 | |
8YXR
 
 | |
9IJ9
 
 | | A Cryo-EM structure of Bitter taste receptor TAS2R14 with Gi complex | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, Q, Duan, J, Tao, L, Xu, E.H. | | Deposit date: | 2024-06-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Bitter taste receptor TAS2R14 activation and G protein assembly by an intracellular agonist.
Cell Res., 2024
|
|
8ZEY
 
 | | Anti-CRISPR type I subtype E3;AcrIE3 | | Descriptor: | AcrIE3 | | Authors: | Kim, D.Y, Park, H.H. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.734 Å) | | Cite: | Novel structure of the anti-CRISPR protein AcrIE3 and its implication on the CRISPR-Cas inhibition.
Biochem.Biophys.Res.Commun., 722, 2024
|
|
8Z9A
 
 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum bound with geranyl acetate | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
8YUA
 
 | | Tubulin-RB3-TTL in complex with compound SI10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloranyl-~{N}-(4-methoxyphenyl)-~{N}-methyl-thieno[3,2-d]pyrimidin-4-amine, CALCIUM ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The complex structure of tubulin-RB3-TTL in complex with compound SI10
To Be Published
|
|
9B8P
 
 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, V1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H+-transporting V1 subunit D, H(+)-transporting two-sector ATPase, ... | | Authors: | Coupland, E.M, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9B55
 
 | |
9B5B
 
 | |
9B5F
 
 | |
6DAT
 
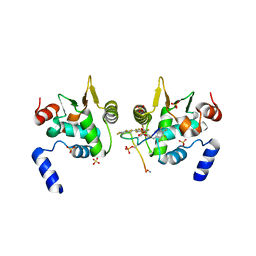 | | ETS1 in complex with synthetic SRR mimic | | Descriptor: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | Authors: | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2018-05-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35002637 Å) | | Cite: | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
9BB1
 
 | |
9BD3
 
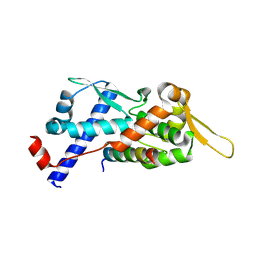 | | Structure of the MAGEA4 MHD-RAD18 R6BD Complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Melanoma antigen A 4 | | Authors: | Forker, K, Zhou, P. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of MAGEA4 MHD-RAD18 R6BD reveals a flipped binding mode compared to AlphaFold2 prediction.
Embo J., 43, 2024
|
|
5KOP
 
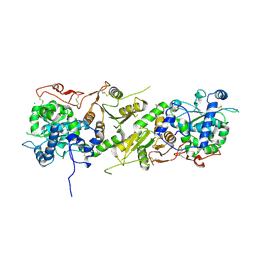 | | Arabidopsis thaliana fucosyltransferase 1 (FUT1) in its apo-form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Galactoside 2-alpha-L-fucosyltransferase | | Authors: | Rocha, J, de Sanctis, D, Breton, C. | | Deposit date: | 2016-07-01 | | Release date: | 2016-10-12 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Arabidopsis thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation.
Plant Cell, 28, 2016
|
|
8XAV
 
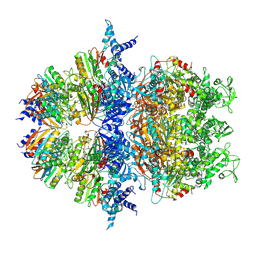 | | Cryo-EM structure of an anti-phage defense complex | | Descriptor: | ATP-binding protein, DUF4297 | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
6DBH
 
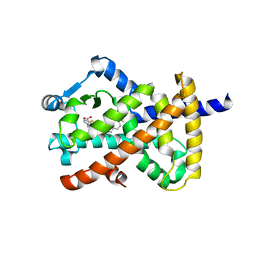 | | Crystal structure of PPAR gamma in complex with NMP422 | | Descriptor: | 4-{2-[(2,3-dioxo-1-pentyl-2,3-dihydro-1H-indol-5-yl)sulfanyl]ethyl}benzoic acid, CHLORIDE ION, Peroxisome proliferator-activated receptor gamma | | Authors: | Mou, T.C, Chrisman, I.M, Hughes, T.S, Sprang, S.R. | | Deposit date: | 2018-05-03 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Crystal structure of PPAR gamma in complex with NMP422
To Be Published
|
|
6DT4
 
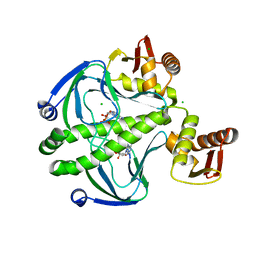 | | 1.8 Angstrom Resolution Crystal Structure of cAMP-Regulatory Protein from Yersinia pestis in Complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CHLORIDE ION, Cyclic AMP receptor protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Ritzert, J.T.H, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-15 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Cyclic AMP Receptor Protein Regulates Quorum Sensing and Global Gene Expression in Yersinia pestis during Planktonic Growth and Growth in Biofilms.
Mbio, 10, 2019
|
|
6DBX
 
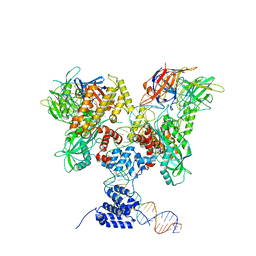 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8ZLV
 
 | |
