5JM0
 
 | | Structure of the S. cerevisiae alpha-mannosidase 1 | | Descriptor: | Alpha-mannosidase,Alpha-mannosidase,Alpha-mannosidase | | Authors: | Schneider, S, Kosinski, J, Jakobi, A.J, Hagen, W.J.H, Sachse, C. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle.
Embo Rep., 17, 2016
|
|
6GIW
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum (Mutation L91P) with Chlorophyll-a | | Descriptor: | CHLOROPHYLL A, Water-soluble chlorophyll protein | | Authors: | Palm, D.M, Agostini, A, Averesch, V, Girr, P, Werwie, M, Takahashi, S, Satoh, H, Jaenicke, E, Paulsen, H. | | Deposit date: | 2018-05-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chlorophyll a/b binding-specificity in water-soluble chlorophyll protein.
Nat Plants, 4, 2018
|
|
5FHZ
 
 | |
8SLY
 
 | | Rat TRPV2 bound with 2 CBD ligands in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 2, ... | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cannabidiol sensitizes TRPV2 channels to activation by 2-APB.
Elife, 12, 2023
|
|
8SLX
 
 | | Rat TRPV2 bound with 1 CBD ligand in nanodiscs | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily V member 2, cannabidiol | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cannabidiol sensitizes TRPV2 channels to activation by 2-APB.
Elife, 12, 2023
|
|
5Z2F
 
 | | NADPH/PDA bound Dihydrodipicolinate reductase from Paenisporosarcina sp. TG-14 | | Descriptor: | Dihydrodipicolinate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Lee, J.H, Lee, C.W, Park, S. | | Deposit date: | 2018-01-02 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dihydrodipicolinate reductase (PaDHDPR) from Paenisporosarcina sp. TG-14: structural basis for NADPH preference as a cofactor
Sci Rep, 8, 2018
|
|
1A4S
 
 | | BETAINE ALDEHYDE DEHYDROGENASE FROM COD LIVER | | Descriptor: | BETAINE ALDEHYDE DEHYDROGENASE | | Authors: | Johansson, K, El Ahmad, M, Hjelmqvist, L, Ramaswamy, S, Jornvall, H, Eklund, H. | | Deposit date: | 1998-02-03 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of betaine aldehyde dehydrogenase at 2.1 A resolution.
Protein Sci., 7, 1998
|
|
4WNI
 
 | |
3LF1
 
 | |
5YX9
 
 | |
5Z2D
 
 | |
2PZH
 
 | | YbgC thioesterase (Hp0496) from Helicobacter pylori | | Descriptor: | Hypothetical protein HP_0496 | | Authors: | Angelini, A, Cendron, L, Goncalves, S, Zanotti, G, Terradot, L. | | Deposit date: | 2007-05-18 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and enzymatic characterization of HP0496, a YbgC thioesterase from Helicobacter pylori.
Proteins, 72, 2008
|
|
5ZMC
 
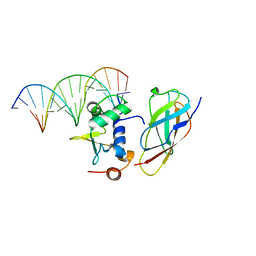 | | Structural Basis for Reactivation of -146C>T Mutant TERT Promoter by cooperative binding of p52 and ETS1/2 | | Descriptor: | DNA (5'-D(P*CP*GP*GP*GP*GP*AP*CP*CP*CP*GP*GP*AP*AP*GP*GP*G)-3'), DNA (5'-D(P*GP*CP*CP*CP*TP*TP*CP*CP*GP*GP*GP*TP*CP*CP*CP*C)-3'), Nuclear factor NF-kappa-B p100 subunit, ... | | Authors: | Xu, X, Bharath, S.R, Song, H. | | Deposit date: | 2018-04-02 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for reactivating the mutant TERT promoter by cooperative binding of p52 and ETS1.
Nat Commun, 9, 2018
|
|
3LF2
 
 | | NADPH Bound Structure of the Short Chain Oxidoreductase Q9HYA2 from Pseudomonas aeruginosa PAO1 Containing an Atypical Catalytic Center | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Huether, R, Umland, T.C, Duax, W.L. | | Deposit date: | 2010-01-15 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The short-chain oxidoreductase Q9HYA2 from Pseudomonas aeruginosa PAO1 contains an atypical catalytic center.
Protein Sci., 19, 2010
|
|
1KYH
 
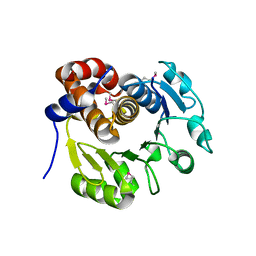 | | Structure of Bacillus subtilis YxkO, a Member of the UPF0031 Family and a Putative Kinase | | Descriptor: | Hypothetical 29.9 kDa protein in SIGY-CYDD intergenic region | | Authors: | Zhang, R, Dementieva, I, Vinokour, E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-04 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Bacillus subtilis YXKO--a member of the UPF0031 family and a putative kinase.
J.Struct.Biol., 139, 2002
|
|
1NNA
 
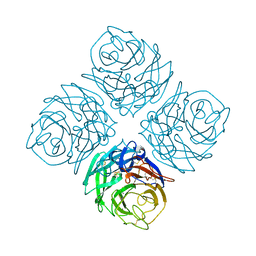 | | THREE-DIMENSIONAL STRUCTURE OF INFLUENZA A N9 NEURAMINIDASE AND ITS COMPLEX WITH THE INHIBITOR 2-DEOXY 2,3-DEHYDRO-N-ACETYL NEURAMINIC ACID | | Descriptor: | CALCIUM ION, NEURAMINIDASE | | Authors: | Bossart-Whitaker, P, Carson, M, Babu, Y.S, Smith, C.D, Laver, W.G, Air, G.M. | | Deposit date: | 1993-03-08 | | Release date: | 1994-04-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid.
J.Mol.Biol., 232, 1993
|
|
6GIX
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum (Mutation L91P) with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-soluble chlorophyll protein | | Authors: | Palm, D.M, Agostini, A, Averesch, V, Girr, P, Werwie, M, Takahashi, S, Satoh, H, Jaenicke, E, Paulsen, H. | | Deposit date: | 2018-05-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chlorophyll a/b binding-specificity in water-soluble chlorophyll protein.
Nat Plants, 4, 2018
|
|
5HIA
 
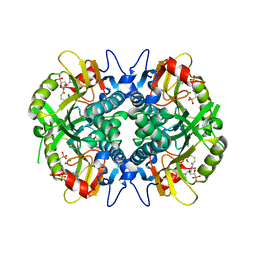 | | Human hypoxanthine-guanine phosphoribosyltransferase in complex with [3R,4R]-4-guanin-9-yl-3-((S)-2-hydroxy-2-phosphonoethyl)oxy-1-N-(phosphonopropionyl)pyrrolidine | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, [3-[(3~{R},4~{R})-3-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-[(2~{S})-2-oxidanyl-2-phosphono-ethoxy]pyrrolidin-1-y l]-3-oxidanylidene-propyl]phosphonic acid | | Authors: | Guddat, L.W, Keough, D.T, Rejman, D. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Design of Plasmodium vivax Hypoxanthine-Guanine Phosphoribosyltransferase Inhibitors as Potential Antimalarial Therapeutics.
ACS Chem. Biol., 2017
|
|
1OJR
 
 | | L-rhamnulose-1-phosphate aldolase from Escherichia coli (mutant E192A) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Dihydroxyacetone, GLYCEROL, ... | | Authors: | Kroemer, M, Merkel, I, Schulz, G.E. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Catalytic Mechanism of L-Rhamnulose-1-Phosphate Aldolase.
Biochemistry, 42, 2003
|
|
2QMX
 
 | | The crystal structure of L-Phe inhibited prephenate dehydratase from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PHENYLALANINE, ... | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
2CNA
 
 | | THE COVALENT AND THREE-DIMENSIONAL STRUCTURE OF CONCANAVALIN A, IV.ATOMIC COORDINATES,HYDROGEN BONDING,AND QUATERNARY STRUCTURE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Reekejunior, G.N, Becker, J.W, Edelman, G.M. | | Deposit date: | 1975-04-01 | | Release date: | 1977-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The covalent and three-dimensional structure of concanavalin A. IV. Atomic coordinates, hydrogen bonding, and quaternary structure.
J.Biol.Chem., 250, 1975
|
|
2QMW
 
 | | The crystal structure of the prephenate dehydratase (PDT) from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Zhang, R, Li, H, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
2L0J
 
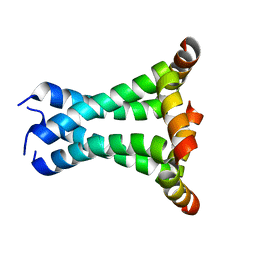 | | Solid State NMR structure of the M2 proton channel from Influenza A Virus in hydrated lipid bilayer | | Descriptor: | Matrix protein 2 | | Authors: | Sharma, M, Yi, M, Dong, H, Qin, H, Peterson, E, Busath, D.D, Zhou, H.X, Cross, T.A. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Insight into the mechanism of the influenza a proton channel from a structure in a lipid bilayer.
Science, 330, 2010
|
|
1QT1
 
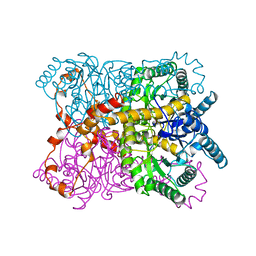 | |
2DTE
 
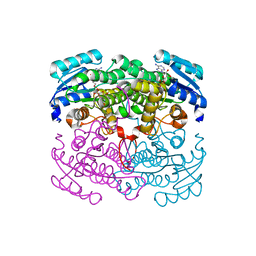 | | Structure of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glucose 1-dehydrogenase related protein | | Authors: | Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | Deposit date: | 2006-07-12 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights into Unique Substrate Selectivity of Thermoplasma acidophilumd-Aldohexose Dehydrogenase
J.Mol.Biol., 367, 2007
|
|
