9J2F
 
 | | Structure of photosynthetic LH1-RC complex from the purple bacterium Blastochloris tepida | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 15-cis-1,2-dihydroneurosporene, Antenna complex alpha/beta subunit domain-containing protein, ... | | Authors: | Kimura, Y, Kanno, R, Mori, K, Matsuda, Y, Seto, R, Takenaka, S, Mino, H, Ohkubo, T, Honda, M, Sasaki, Y.C, Kishikawa, J, Mitsuoka, K, Mio, K, Hall, M, Purba, E.R, Mochizuki, T, Mizoguchi, A, Humbel, B.M, Madigan, M.T, Wang-Otomo, Z.-Y, Tani, K. | | Deposit date: | 2024-08-06 | | Release date: | 2024-12-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | The Thermal-Stable LH1-RC Complex of a Hot Spring Purple Bacterium Powers Photosynthesis with Extremely Low-Energy Near-Infrared Light.
Biochemistry, 64, 2025
|
|
6Y2L
 
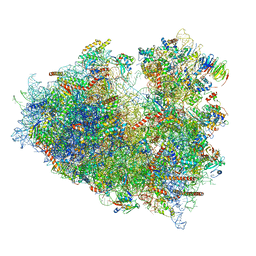 | | Structure of human ribosome in POST state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-16 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
7WVF
 
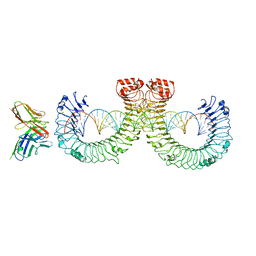 | | ectoTLR3-mAb12-poly(I:C) complex | | Descriptor: | RNA (46-MER), Toll-like receptor 3, mAb12 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7X6Z
 
 | |
8QW1
 
 | | Human NDPK-C in complex with ADP and Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase 3 | | Authors: | Werten, S, Amjadi, R, Scheffzek, K. | | Deposit date: | 2023-10-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic Insights into Substrate Recognition of Human Nucleoside Diphosphate Kinase C Based on Nucleotide-Induced Structural Changes.
Int J Mol Sci, 25, 2024
|
|
7X70
 
 | |
4Y8D
 
 | | Crystal structure of Cyclin-G associated kinase (GAK) complexed with selective 12i inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-[3-(morpholin-4-yl)[1,2]thiazolo[4,3-b]pyridin-6-yl]aniline, Cyclin-G-associated kinase, ... | | Authors: | Chaikuad, A, Heroven, C, Nowak, R, De Jonghe, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective Inhibitors of Cyclin G Associated Kinase (GAK) as Anti-Hepatitis C Agents.
J.Med.Chem., 58, 2015
|
|
8QVY
 
 | | Human NDPK-C unliganded | | Descriptor: | Nucleoside diphosphate kinase 3, PHOSPHATE ION | | Authors: | Werten, S, Amjadi, R, Scheffzek, K. | | Deposit date: | 2023-10-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Mechanistic Insights into Substrate Recognition of Human Nucleoside Diphosphate Kinase C Based on Nucleotide-Induced Structural Changes.
Int J Mol Sci, 25, 2024
|
|
8QW2
 
 | | Human NDPK-C in complex with UDP and Mg2+ | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase 3, URIDINE-5'-DIPHOSPHATE | | Authors: | Werten, S, Amjadi, R, Scheffzek, K. | | Deposit date: | 2023-10-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanistic Insights into Substrate Recognition of Human Nucleoside Diphosphate Kinase C Based on Nucleotide-Induced Structural Changes.
Int J Mol Sci, 25, 2024
|
|
7X6Y
 
 | |
9F2I
 
 | | Crystal structure of SARS-CoV-2 N-protein C-terminal domain in complex with 2-amino-1,3-benzothiazol-6-ol | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-1,3-benzothiazol-6-ol, Nucleoprotein, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B, Perez-Canadillas, J.M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The ALS drug riluzole binds to the C-terminal domain of SARS-CoV-2 nucleocapsid protein and has antiviral activity.
Structure, 33, 2025
|
|
6VVG
 
 | | Structure of the Cydia pomonella Granulovirus kinase, PK-1 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Arginine kinase | | Authors: | Oliver, M.R, Horne, C.R, Keown, J.R, Murphy, J.M, Metcalf, P. | | Deposit date: | 2020-02-17 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Granulovirus PK-1 kinase activity relies on a side-to-side dimerization mode centered on the regulatory alpha C helix.
Nat Commun, 12, 2021
|
|
4WHI
 
 | | Crystal structure of C-terminal domain of penicillin binding protein Rv0907 | | Descriptor: | BROMIDE ION, Beta-lactamase, NICKEL (II) ION | | Authors: | Chang, C, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-22 | | Release date: | 2014-10-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of C-terminal domain of penicillin binding protein Rv0907
To Be Published
|
|
8R0T
 
 | |
6QNV
 
 | | Fibrinogen-like globe domain of Human Tenascin-C | | Descriptor: | Tenascin | | Authors: | Coker, J.A, Bezerra, G.A, Bradshaw, W.J, Zhang, M, Yosaatmadja, Y, Fernandez-Cid, A, Shrestha, L, Burgess-Brown, N, Gileadi, O, Arrowsmith, C.H, Bountra, C, Midwood, K.S, Yue, W.W, Marsden, B.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fibrinogen-like globe domain of Human Tenascin-C
To Be Published
|
|
6DIU
 
 | | Crystal structure of HCV NS3/4A protease in complex with P4-3(AJ-74) | | Descriptor: | 1-methylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, GLYCEROL, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Design of Hepatitis C NS3/4A Protease Inhibitors Leveraging Untapped Regions of the Substrate Envelope
To Be Published
|
|
6DIR
 
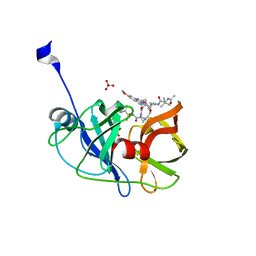 | | Crystal structure of HCV NS3/4A protease in complex with P4-P5-2 (AJ-67) | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(N-acetyl-3-methyl-L-valyl)amino]-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-N-[(1-methyl cyclopropyl)sulfonyl]-5,16-dioxo-1,2,3,6,7,8,9,10,11,13a,14,15,16,16a-tetradecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diaz acyclopentadecine-14a(5H)-carboxamide, NS3 protease, SULFATE ION, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Design of Hepatitis C NS3/4A Protease Inhibitors Leveraging Untapped Regions of the Substrate Envelope
To Be Published
|
|
6DIV
 
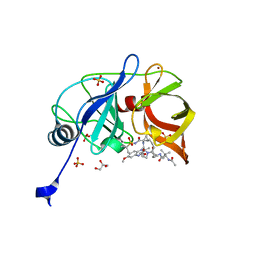 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-P5-2 (AJ-67) | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(N-acetyl-3-methyl-L-valyl)amino]-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-N-[(1-methyl cyclopropyl)sulfonyl]-5,16-dioxo-1,2,3,6,7,8,9,10,11,13a,14,15,16,16a-tetradecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diaz acyclopentadecine-14a(5H)-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design of Hepatitis C NS3/4A Protease Inhibitors Leveraging Untapped Regions of the Substrate Envelope
To Be Published
|
|
6DIT
 
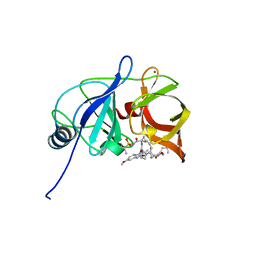 | | Crystal structure of HCV NS3/4A protease in complex with P4-2 (JZ01-19) | | Descriptor: | NS3 protease, ZINC ION, pentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Design of Hepatitis C NS3/4A Protease Inhibitors Leveraging Untapped Regions of the Substrate Envelope
To Be Published
|
|
6T41
 
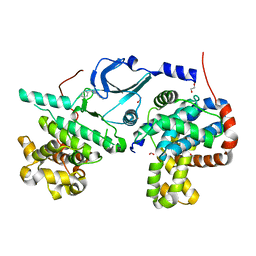 | | CDK8/Cyclin C in complex with N-(4-chlorobenzyl)isoquinolin-4-amine | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Schneider, E.V, Maskos, K, Huber, R, Kuhn, C.-D. | | Deposit date: | 2019-10-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A precisely positioned MED12 activation helix stimulates CDK8 kinase activity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7N1Z
 
 | | NMR structure of native PnIA | | Descriptor: | Alpha-conotoxin PnIA | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N24
 
 | | NMR structure of native EpI | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N25
 
 | | NMR structure of EpI-OH | | Descriptor: | Alpha-conotoxin EpI-OH | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N26
 
 | | NMR structure of EpI-[Y(SO3)15Y]-NH2 | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
8A8X
 
 | | TRIM7 PRYSPRY in complex with a MNV1-NS3 peptide HDDFGLQ | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, MNV1-NS3 C term peptide | | Authors: | Luptak, J. | | Deposit date: | 2022-06-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | TRIM7 Restricts Coxsackievirus and Norovirus Infection by Detecting the C-Terminal Glutamine Generated by 3C Protease Processing.
Viruses, 14, 2022
|
|
