1TVI
 
 | | Solution structure of TM1509 from Thermotoga maritima: VT1, a NESGC target protein | | Descriptor: | Hypothetical UPF0054 protein TM1509 | | Authors: | Penhoat, C.H, Atreya, H.S, Kim, S, Li, Z, Yee, A, Xiao, R, Murray, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Thermotoga maritima protein TM1509 reveals a Zn-metalloprotease-like tertiary structure.
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
2OFG
 
 | | Solution structure of the n-terminal domain of the zinc(II) ATPase ziaa in its apo form | | Descriptor: | Zinc-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Poggi, L, Robinson, N.J, Vanarotti, M. | | Deposit date: | 2007-01-03 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of the soluble domain of ZiaA-ATPase and the basis of selective interactions with copper metallochaperone Atx1.
J.Biol.Inorg.Chem., 15, 2010
|
|
1ROQ
 
 | |
2OFH
 
 | | Solution structure of the n-terminal domain of the zinc(II) ATPase ziaa in its apo form | | Descriptor: | Zinc-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Poggi, L, Robinson, N.J, Vanarotti, M. | | Deposit date: | 2007-01-03 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of the soluble domain of ZiaA-ATPase and the basis of selective interactions with copper metallochaperone Atx1.
J.Biol.Inorg.Chem., 15, 2010
|
|
1ZRP
 
 | | SOLUTION-STATE STRUCTURE BY NMR OF ZINC-SUBSTITUTED RUBREDOXIN FROM THE MARINE HYPERTHERMOPHILIC ARCHAEBACTERIUM PYROCOCCUS FURIOSUS | | Descriptor: | RUBREDOXIN, ZINC ION | | Authors: | Blake, P.R, Park, J.B, Zhou, Z.H, Hare, D.R, Adams, M.W.W, Summers, M.F. | | Deposit date: | 1992-07-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution-state structure by NMR of zinc-substituted rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus.
Protein Sci., 1, 1992
|
|
1V66
 
 | | Solution structure of human p53 binding domain of PIAS-1 | | Descriptor: | Protein inhibitor of activated STAT protein 1 | | Authors: | Okubo, S, Hara, F, Tsuchida, Y, Shimotakahara, S, Suzuki, S, Hatanaka, H, Yokoyama, S, Tanaka, H, Yasuda, H, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-12-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers
J.Biol.Chem., 279, 2004
|
|
2JM4
 
 | |
1IMI
 
 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 | | Descriptor: | PROTEIN (ALPHA-CONOTOXIN IMI) | | Authors: | Maslennikov, I.V, Shenkarev, Z.O, Zhmak, M.N, Tsetlin, V.I, Ivanov, V.T, Arseniev, A.S. | | Deposit date: | 1998-11-27 | | Release date: | 1999-04-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR spatial structure of alpha-conotoxin ImI reveals a common scaffold in snail and snake toxins recognizing neuronal nicotinic acetylcholine receptors.
FEBS Lett., 444, 1999
|
|
2KNR
 
 | | Solution structure of protein Atu0922 from A. tumefaciens. Northeast Structural Genomics Consortium target AtT13. Ontario Center for Structural Proteomics target ATC0905 | | Descriptor: | Uncharacterized protein ATC0905 | | Authors: | Gutmanas, A, Yee, S, Lemak, A, Fares, A, Semesi, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein atc0905 from A. tumefaciens
To be Published
|
|
1J3G
 
 | | Solution structure of Citrobacter Freundii AmpD | | Descriptor: | AmpD protein, ZINC ION | | Authors: | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
1YFV
 
 | |
2HZ8
 
 | | QM/MM structure refined from NMR-structure of a single chain diiron protein | | Descriptor: | De novo designed diiron protein, ZINC ION | | Authors: | Calhoun, J.R, Liu, W, Spiegel, K, Dal Peraro, M, Klein, M.L, Wand, A.J, DeGrado, W.F. | | Deposit date: | 2006-08-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement.
Structure, 16, 2008
|
|
2DCR
 
 | |
2JSV
 
 | | Dipole tensor-based refinement for atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR spectroscopy | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Franks, W, Wylie, B.J, Frericks, H.L, Nieuwkoop, A.J, Mayrhofer, R, Shah, G.J, Graesser, D.T, Rienstra, C.M. | | Deposit date: | 2007-07-16 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-29 | | Method: | SOLID-STATE NMR | | Cite: | Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2KJ3
 
 | | High-resolution structure of the HET-s(218-289) prion in its amyloid form obtained by solid-state NMR | | Descriptor: | Small s protein | | Authors: | Van Melckebeke, H, Wasmer, C, Lange, A, AB, E, Loquet, A, Meier, B.H. | | Deposit date: | 2009-05-20 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of HET-s(218-289) Amyloid Fibrils by Solid-State NMR Spectroscopy
J.Am.Chem.Soc., 132, 2010
|
|
2CZP
 
 | | Structural analysis of membrane-bound mastoparan-X by solid-state NMR | | Descriptor: | Mastoparan X | | Authors: | Todokoro, Y, Fujiwara, T, Yumen, I, Fukushima, K, Kang, S.-W, Park, J.-S, Kohno, T, Wakamatsu, K, Akutsu, H. | | Deposit date: | 2005-07-14 | | Release date: | 2006-07-04 | | Last modified: | 2022-03-09 | | Method: | SOLID-STATE NMR | | Cite: | Structure of tightly membrane-bound mastoparan-x, a g-protein-activating Peptide, determined by solid-state NMR.
Biophys.J., 91, 2006
|
|
1ZNT
 
 | | 18 NMR structures of AcAMP2-Like Peptide with non Natural Fluoroaromatic Residue (AcAMP2F18Pff/Y20Pff) complex with N,N,N-triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Aboitiz, N, Freire, F, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-05-12 | | Release date: | 2005-12-06 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues
Chemistry, 11, 2005
|
|
1ZUV
 
 | | 24 NMR structures of AcAMP2-Like Peptide with Phenylalanine 18 mutated to Tryptophan | | Descriptor: | AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Freire, F, Aboitiz, N, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-06-01 | | Release date: | 2005-12-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues
Chemistry, 11, 2005
|
|
2KP6
 
 | | Solution NMR structure of protein CV0237 from Chromobacterium violaceum. Northeast Structural Genomics Consortium (NESG) target CvT1 | | Descriptor: | Uncharacterized protein | | Authors: | Fares, C, Lemak, A, Yee, A, Garcia, M, Montelione, G.T, Arrowsmith, C.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein CV0237 from Chromobacterium violaceum
TO BE PUBLISHED
|
|
1ZWU
 
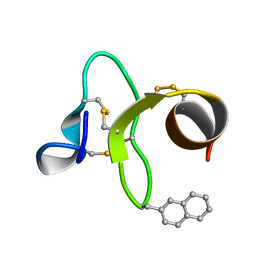 | | 30 NMR structures of AcAMP2-like peptide with non natural beta-(2-naphthyl)-alanine residue. | | Descriptor: | AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 (ACMP2) | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Freire, F, Aboitiz, N, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-06-06 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues.
Chemistry, 11, 2005
|
|
2MPZ
 
 | |
2ECH
 
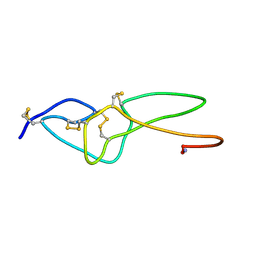 | |
2FLY
 
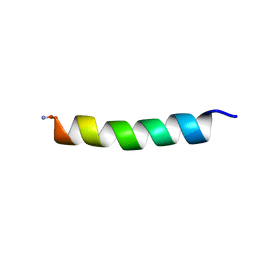 | | Proadrenomedullin N-Terminal 20 Peptide | | Descriptor: | Proadrenomedullin N-20 terminal peptide | | Authors: | Lucyk, S, Taha, H, Yamamoto, H, Miskolzie, M, Kotovych, G. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR conformational analysis of proadrenomedullin N-terminal 20 peptide, a proangiogenic factor involved in tumor growth
Biopolymers, 81, 2006
|
|
2JN4
 
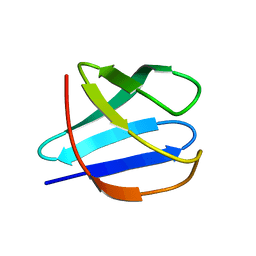 | | Solution NMR Structure of Protein RP4601 from Rhodopseudomonas palustris. Northeast Structural Genomics Consortium Target RpT2; Ontario Center for Structural Proteomics Target RP4601. | | Descriptor: | Hypothetical protein fixU, nifT | | Authors: | Lemak, A, Srisailam, S, Yee, A, Karra, M.D, Lukin, J.A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-22 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hypothetical protein RP4601 from Rhodopseudomonas palustris
To be Published
|
|
2K54
 
 | | Solution NMR structure of protein Atu0742 from Agrobacterium Tumefaciens. Northeast Structural Genomics Consortium (NESG0) target AtT8. Ontario Center for Structural Proteomics target ATC0727 . | | Descriptor: | Protein Atu0742 | | Authors: | Lemak, A, Yee, A, Gutmanas, A, Fares, C, Semesi, A, Arrowsmith, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein ATC0727 from Agrobacterium Tumefaciens.
To be Published
|
|
